Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC018462A_C01 KMC018462A_c01
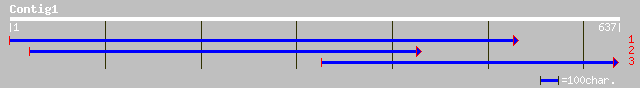
(637 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAD18921.1| RNA-binding protein precursor [Persea americana] 151 8e-36
pir||T06817 RNA-binding protein - garden pea gi|2330647|emb|CAA7... 148 5e-35
pir||S50765 RNA-binding protein - common ice plant gi|388621|gb|... 134 8e-31
ref|NP_194208.1| 31 kDa ribonucleoprotein, chloroplast precursor... 131 9e-30
gb|AAK95304.1|AF410318_1 AT4g24770/F22K18_30 [Arabidopsis thalia... 131 9e-30
>emb|CAD18921.1| RNA-binding protein precursor [Persea americana]
Length = 300
Score = 151 bits (381), Expect = 8e-36
Identities = 97/171 (56%), Positives = 114/171 (65%), Gaps = 8/171 (4%)
Frame = +2
Query: 149 PSS--LFYNKTKHLFFSAPSRPLKLQLSSINPSPALSLAATTHHRS-SAPLFVAQTSDWA 319
PSS LF +K F PS+P+KL S + S +S + S S VAQTSDWA
Sbjct: 3 PSSPILFSSKPPCPFLPIPSKPIKLIHLSTSSSSWVSFKKSPSPSSRSLTTLVAQTSDWA 62
Query: 320 QQEEDNTL---TLEDQE-EG-AEGVSETEAGWEPNGEDAEIEGAEEGEGEEGDYEEPPEE 484
QQEED+ + TL D E EG EG +TE+G E +GE++ +G EG GEE Y PPEE
Sbjct: 63 QQEEDSNVAEATLSDWEGEGEGEGEEQTESGAE-DGEESGGDGFVEG-GEEDSYPLPPEE 120
Query: 485 AKLFVGNLPYDADGHKLAELFNQVGTVDLAEVIYNRETDQSRGFGFVTMKT 637
AKLFVGNLPYD D LAELF+Q GTV++AEVIYNRETDQSRGFGFVTM T
Sbjct: 121 AKLFVGNLPYDVDHQALAELFDQAGTVEVAEVIYNRETDQSRGFGFVTMST 171
Score = 59.3 bits (142), Expect = 4e-08
Identities = 26/50 (52%), Positives = 38/50 (76%)
Frame = +2
Query: 488 KLFVGNLPYDADGHKLAELFNQVGTVDLAEVIYNRETDQSRGFGFVTMKT 637
+++VGNLP+ D +L ++F++ G V A V+Y+RET +SRGFGFVTM T
Sbjct: 216 RMYVGNLPWQVDDARLEQVFSEHGKVVEARVVYDRETGRSRGFGFVTMST 265
>pir||T06817 RNA-binding protein - garden pea gi|2330647|emb|CAA74889.1|
ribonucleoprotein [Pisum sativum]
gi|10179830|gb|AAG13900.1| 33 kDa ribonucleoprotein
[Pisum sativum]
Length = 291
Score = 148 bits (374), Expect = 5e-35
Identities = 98/189 (51%), Positives = 114/189 (59%), Gaps = 6/189 (3%)
Frame = +2
Query: 89 MSITS----FKSLTTMAESCLLSAPSSLFYNKTKHLFFSAPSRPLKLQLSSINPSPALSL 256
MS+TS FKSLT A LF+NKTK FS PS+P K +S +N SP+L+L
Sbjct: 1 MSVTSTTPLFKSLTIAA----------LFHNKTKPTSFSLPSKPFKFHIS-LNSSPSLTL 49
Query: 257 AATTHHRSSAPLFVAQTSDWAQQEEDNTLTLEDQEEGAEGVSETEA--GWEPNGEDAEIE 430
+ T+ + PLF AQ E TLT E EGV ETE WEP + E
Sbjct: 50 SLKTNR--ATPLFAAQ--------EGETLTTE------EGVVETEGLIDWEPEAAENET- 92
Query: 431 GAEEGEGEEGDYEEPPEEAKLFVGNLPYDADGHKLAELFNQVGTVDLAEVIYNRETDQSR 610
G E+ G GD+ EP E+AKLFVGNLPYD D KLA LF GTV++AEVIYNRETDQSR
Sbjct: 93 GGEDYAG--GDFAEPSEDAKLFVGNLPYDVDSEKLAMLFEPAGTVEIAEVIYNRETDQSR 150
Query: 611 GFGFVTMKT 637
GFGFVTM T
Sbjct: 151 GFGFVTMST 159
Score = 59.7 bits (143), Expect = 3e-08
Identities = 35/100 (35%), Positives = 54/100 (54%), Gaps = 4/100 (4%)
Frame = +2
Query: 344 TLEDQEEGAEGVSETEAGWEPNGEDAEIEGAEEGEGEEGDYEEPPEE----AKLFVGNLP 511
T+E+ E GA + ++ NG + A E PP +++VGNL
Sbjct: 159 TVEEAEAGAAKFNR----YDYNGRPLTVNKAAPRGSRPEREERPPRTFEPVLRVYVGNLS 214
Query: 512 YDADGHKLAELFNQVGTVDLAEVIYNRETDQSRGFGFVTM 631
++ D +L ++F++ G V A V+Y+RET +SRGFGFVTM
Sbjct: 215 WELDDSRLEQVFSEHGKVVSARVVYDRETGRSRGFGFVTM 254
>pir||S50765 RNA-binding protein - common ice plant gi|388621|gb|AAA33039.1|
RNA-binding protein
Length = 289
Score = 134 bits (338), Expect = 8e-31
Identities = 80/174 (45%), Positives = 104/174 (58%), Gaps = 2/174 (1%)
Frame = +2
Query: 122 MAESCLLSAPSSLFYNKTKHLFFSAPSRPLKLQLSSINPSPALSLAATTHHRSSAPL--F 295
M+ +C S+ S K LFFS PS+P K+ LS SP LS + S+ + +
Sbjct: 1 MSNTCFTSSLHSPSILIPKSLFFSIPSKPTKIPLSLSLSSPQLSSFSLQKQTLSSTVVTY 60
Query: 296 VAQTSDWAQQEEDNTLTLEDQEEGAEGVSETEAGWEPNGEDAEIEGAEEGEGEEGDYEEP 475
VAQTS+W Q+ + L EEG S+ E W G++EGEG ++EP
Sbjct: 61 VAQTSEWDQEGTNAVL-----EEG----SDQEPSWGNTEAQVSDFGSDEGEG----FQEP 107
Query: 476 PEEAKLFVGNLPYDADGHKLAELFNQVGTVDLAEVIYNRETDQSRGFGFVTMKT 637
PE+AKLFVGNLP+D D KLA++F G V++AEVIYNRETD+SRGFGFVTM T
Sbjct: 108 PEDAKLFVGNLPFDVDSEKLAQIFEGAGVVEIAEVIYNRETDRSRGFGFVTMST 161
Score = 57.8 bits (138), Expect = 1e-07
Identities = 35/91 (38%), Positives = 50/91 (54%), Gaps = 1/91 (1%)
Frame = +2
Query: 362 EGAEGVSETEAGWEPNGEDAEI-EGAEEGEGEEGDYEEPPEEAKLFVGNLPYDADGHKLA 538
E AE E +E NG + + A G E E +++VGNLP+D D +L
Sbjct: 163 EEAEKAVELYHKFEVNGRFLTVNKAAPRGSRPERAPREYEPSFRVYVGNLPWDVDDARLE 222
Query: 539 ELFNQVGTVDLAEVIYNRETDQSRGFGFVTM 631
++F++ G V A V+ +RET +SRGF FVTM
Sbjct: 223 QVFSEHGKVLSARVVSDRETGRSRGFAFVTM 253
>ref|NP_194208.1| 31 kDa ribonucleoprotein, chloroplast precursor (RNA-binding
protein cp31); protein id: At4g24770.1, supported by
cDNA: gi_15294253, supported by cDNA: gi_387568
[Arabidopsis thaliana] gi|464662|sp|Q04836|ROC3_ARATH 31
kDa ribonucleoprotein, chloroplast precursor
(RNA-binding protein RNP-T) (RNA-binding protein 1/2/3)
(AtRBP33) (RNA-binding protein cp31)
gi|282884|pir||S28057 RNA-binding protein RNP-T
precursor - Arabidopsis thaliana
gi|16490|emb|CAA46347.1| RNA-binding protein
[Arabidopsis thaliana] gi|387569|gb|AAA32860.1| 31 kDa
RNA binding protein gi|475718|gb|AAA18378.1| RNA-binding
protein 1 gi|4220513|emb|CAA22986.1| RNA-binding protein
RNP-T precursor [Arabidopsis thaliana]
gi|7269328|emb|CAB79387.1| RNA-binding protein RNP-T
precursor [Arabidopsis thaliana] gi|737169|prf||1921382A
RNA-binding protein
Length = 329
Score = 131 bits (329), Expect = 9e-30
Identities = 87/213 (40%), Positives = 117/213 (54%), Gaps = 10/213 (4%)
Frame = +2
Query: 29 IPSSFTPLLFSPSSSSSTSSMSITSFKSLTTMAESCLLSAPSSLFYNKTKHLFFSAPSRP 208
+ SS PL + SSSS+ S S +T++ S + S+ SL +
Sbjct: 6 VTSSLKPLAMADSSSSTIFSHPSIS----STISSSRIRSSSVSLLTGRIN---------- 51
Query: 209 LKLQLSSINPSPALSLAATTHHRSSAPL-FVAQTSDWAQQ---------EEDNTLTLEDQ 358
L L S ++ LSL TH + S + FVAQTSDWA++ E +N+L +D
Sbjct: 52 LPLSFSRVS----LSLKTKTHLKKSPFVSFVAQTSDWAEEGGEGSVAVEETENSLESQDV 107
Query: 359 EEGAEGVSETEAGWEPNGEDAEIEGAEEGEGEEGDYEEPPEEAKLFVGNLPYDADGHKLA 538
EG E + G G+++E + +E E ++ EP EEAKLFVGNL YD + LA
Sbjct: 108 SEGDESEGDASEGDVSEGDESEGDVSEGAVSERAEFPEPSEEAKLFVGNLAYDVNSQALA 167
Query: 539 ELFNQVGTVDLAEVIYNRETDQSRGFGFVTMKT 637
LF Q GTV++AEVIYNRETDQSRGFGFVTM +
Sbjct: 168 MLFEQAGTVEIAEVIYNRETDQSRGFGFVTMSS 200
Score = 60.8 bits (146), Expect = 1e-08
Identities = 27/48 (56%), Positives = 38/48 (78%)
Frame = +2
Query: 488 KLFVGNLPYDADGHKLAELFNQVGTVDLAEVIYNRETDQSRGFGFVTM 631
+++VGNLP+D D +L +LF++ G V A V+Y+RET +SRGFGFVTM
Sbjct: 245 RVYVGNLPWDVDNGRLEQLFSEHGKVVEARVVYDRETGRSRGFGFVTM 292
>gb|AAK95304.1|AF410318_1 AT4g24770/F22K18_30 [Arabidopsis thaliana]
gi|23505889|gb|AAN28804.1| At4g24770/F22K18_30
[Arabidopsis thaliana]
Length = 329
Score = 131 bits (329), Expect = 9e-30
Identities = 87/213 (40%), Positives = 117/213 (54%), Gaps = 10/213 (4%)
Frame = +2
Query: 29 IPSSFTPLLFSPSSSSSTSSMSITSFKSLTTMAESCLLSAPSSLFYNKTKHLFFSAPSRP 208
+ SS PL + SSSS+ S S +T++ S + S+ SL +
Sbjct: 6 VTSSLKPLAMADSSSSTIFSHPSIS----STISSSRIRSSSVSLLTGRIN---------- 51
Query: 209 LKLQLSSINPSPALSLAATTHHRSSAPL-FVAQTSDWAQQ---------EEDNTLTLEDQ 358
L L S ++ LSL TH + S + FVAQTSDWA++ E +N+L +D
Sbjct: 52 LPLSFSRVS----LSLKTKTHLKKSPFVSFVAQTSDWAEEGGEGSVAVEETENSLESQDV 107
Query: 359 EEGAEGVSETEAGWEPNGEDAEIEGAEEGEGEEGDYEEPPEEAKLFVGNLPYDADGHKLA 538
EG E + G G+++E + +E E ++ EP EEAKLFVGNL YD + LA
Sbjct: 108 SEGDESEGDASEGDVSEGDESEGDVSEGAVSERAEFPEPSEEAKLFVGNLAYDVNSQALA 167
Query: 539 ELFNQVGTVDLAEVIYNRETDQSRGFGFVTMKT 637
LF Q GTV++AEVIYNRETDQSRGFGFVTM +
Sbjct: 168 MLFEQAGTVEIAEVIYNRETDQSRGFGFVTMSS 200
Score = 60.8 bits (146), Expect = 1e-08
Identities = 27/48 (56%), Positives = 38/48 (78%)
Frame = +2
Query: 488 KLFVGNLPYDADGHKLAELFNQVGTVDLAEVIYNRETDQSRGFGFVTM 631
+++VGNLP+D D +L +LF++ G V A V+Y+RET +SRGFGFVTM
Sbjct: 245 RVYVGNLPWDVDNGRLEQLFSEHGKVVEARVVYDRETGRSRGFGFVTM 292
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 647,745,348
Number of Sequences: 1393205
Number of extensions: 17498176
Number of successful extensions: 243099
Number of sequences better than 10.0: 7070
Number of HSP's better than 10.0 without gapping: 128323
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 208748
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 26439068301
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)