Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC018228A_C01 KMC018228A_c01
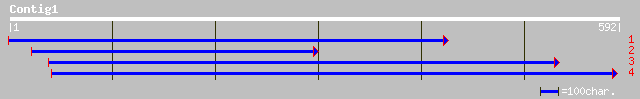
(587 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_565585.1| expressed protein; protein id: At2g25110.1, sup... 195 4e-49
pir||D84644 hypothetical protein At2g25110 [imported] - Arabidop... 195 4e-49
gb|AAM65625.1| unknown [Arabidopsis thaliana] 190 1e-47
gb|EAA00948.1| agCP12010 [Anopheles gambiae str. PEST] 85 8e-16
ref|NP_649527.1| CG11999-PA [Drosophila melanogaster] gi|7296765... 83 2e-15
>ref|NP_565585.1| expressed protein; protein id: At2g25110.1, supported by cDNA:
40987., supported by cDNA: gi_16209644 [Arabidopsis
thaliana] gi|24212393|sp|Q93ZE8|SDF2_ARATH Stromal
cell-derived factor 2-like protein precursor (SDF2-like
protein) gi|16209645|gb|AAL14383.1| At2g25110/F13D4.70
[Arabidopsis thaliana] gi|23505803|gb|AAN28761.1|
At2g25110/F13D4.70 [Arabidopsis thaliana]
Length = 218
Score = 195 bits (495), Expect = 4e-49
Identities = 85/102 (83%), Positives = 95/102 (92%)
Frame = -1
Query: 587 WLHSHLHASPISGNLEVSCFGGESESDTGDYWKVFIEGSGKTWKQDQKIRLQHIDTGGYL 408
WLHSHLHASPISGNLEVSCFG ++ SDTGD+WK+ IEGSGKTWKQDQ++RLQHIDT GYL
Sbjct: 115 WLHSHLHASPISGNLEVSCFGDDTNSDTGDHWKLIIEGSGKTWKQDQRVRLQHIDTSGYL 174
Query: 407 HSHDKKYSRIAGGQQEVCAVREKRADNVWLAAEGVYLPVTES 282
HSHDKKY RIAGGQQEVC +REK+ADN+WLAAEGVYLP+ ES
Sbjct: 175 HSHDKKYQRIAGGQQEVCGIREKKADNIWLAAEGVYLPLNES 216
Score = 35.0 bits (79), Expect = 0.70
Identities = 29/88 (32%), Positives = 38/88 (42%), Gaps = 6/88 (6%)
Frame = -1
Query: 575 HLHASPI-SGNLEVSCFGGESESDTGDYWKV-----FIEGSGKTWKQDQKIRLQHIDTGG 414
H H P SG+ + S G D+ YW V E G K IRLQH+ T
Sbjct: 55 HSHDVPYGSGSGQQSVTGFPGVVDSNSYWIVKPVPGTTEKQGDAVKSGATIRLQHMKTRK 114
Query: 413 YLHSHDKKYSRIAGGQQEVCAVREKRAD 330
+LHSH S I+G + C + +D
Sbjct: 115 WLHSH-LHASPISGNLEVSCFGDDTNSD 141
>pir||D84644 hypothetical protein At2g25110 [imported] - Arabidopsis thaliana
Length = 173
Score = 195 bits (495), Expect = 4e-49
Identities = 85/102 (83%), Positives = 95/102 (92%)
Frame = -1
Query: 587 WLHSHLHASPISGNLEVSCFGGESESDTGDYWKVFIEGSGKTWKQDQKIRLQHIDTGGYL 408
WLHSHLHASPISGNLEVSCFG ++ SDTGD+WK+ IEGSGKTWKQDQ++RLQHIDT GYL
Sbjct: 70 WLHSHLHASPISGNLEVSCFGDDTNSDTGDHWKLIIEGSGKTWKQDQRVRLQHIDTSGYL 129
Query: 407 HSHDKKYSRIAGGQQEVCAVREKRADNVWLAAEGVYLPVTES 282
HSHDKKY RIAGGQQEVC +REK+ADN+WLAAEGVYLP+ ES
Sbjct: 130 HSHDKKYQRIAGGQQEVCGIREKKADNIWLAAEGVYLPLNES 171
Score = 35.0 bits (79), Expect = 0.70
Identities = 29/88 (32%), Positives = 38/88 (42%), Gaps = 6/88 (6%)
Frame = -1
Query: 575 HLHASPI-SGNLEVSCFGGESESDTGDYWKV-----FIEGSGKTWKQDQKIRLQHIDTGG 414
H H P SG+ + S G D+ YW V E G K IRLQH+ T
Sbjct: 10 HSHDVPYGSGSGQQSVTGFPGVVDSNSYWIVKPVPGTTEKQGDAVKSGATIRLQHMKTRK 69
Query: 413 YLHSHDKKYSRIAGGQQEVCAVREKRAD 330
+LHSH S I+G + C + +D
Sbjct: 70 WLHSH-LHASPISGNLEVSCFGDDTNSD 96
>gb|AAM65625.1| unknown [Arabidopsis thaliana]
Length = 218
Score = 190 bits (482), Expect = 1e-47
Identities = 84/102 (82%), Positives = 94/102 (91%)
Frame = -1
Query: 587 WLHSHLHASPISGNLEVSCFGGESESDTGDYWKVFIEGSGKTWKQDQKIRLQHIDTGGYL 408
WLHSHLHASPISGNLEVSCFG ++ SDTGD+WK+ IEGSGKTWKQDQ++RLQHIDT GYL
Sbjct: 115 WLHSHLHASPISGNLEVSCFGDDTNSDTGDHWKLIIEGSGKTWKQDQRVRLQHIDTSGYL 174
Query: 407 HSHDKKYSRIAGGQQEVCAVREKRADNVWLAAEGVYLPVTES 282
HSHDKKY RIAGGQQEVC +REK+ADN+ LAAEGVYLP+ ES
Sbjct: 175 HSHDKKYQRIAGGQQEVCGIREKKADNICLAAEGVYLPLNES 216
Score = 35.0 bits (79), Expect = 0.70
Identities = 29/88 (32%), Positives = 38/88 (42%), Gaps = 6/88 (6%)
Frame = -1
Query: 575 HLHASPI-SGNLEVSCFGGESESDTGDYWKV-----FIEGSGKTWKQDQKIRLQHIDTGG 414
H H P SG+ + S G D+ YW V E G K IRLQH+ T
Sbjct: 55 HSHDVPYGSGSGQQSVTGFPGVVDSNSYWIVKPVPGTTEKQGDAVKSGATIRLQHMKTRK 114
Query: 413 YLHSHDKKYSRIAGGQQEVCAVREKRAD 330
+LHSH S I+G + C + +D
Sbjct: 115 WLHSH-LHASPISGNLEVSCFGDDTNSD 141
>gb|EAA00948.1| agCP12010 [Anopheles gambiae str. PEST]
Length = 231
Score = 84.7 bits (208), Expect = 8e-16
Identities = 44/101 (43%), Positives = 59/101 (57%), Gaps = 1/101 (0%)
Frame = -1
Query: 584 LHSHLHASPISGNLEVSCFGGE-SESDTGDYWKVFIEGSGKTWKQDQKIRLQHIDTGGYL 408
LHSH SP+SGN E+S +G E E DTGD+W V SG +W + +RLQHIDT YL
Sbjct: 122 LHSHHFQSPLSGNQEISAYGDEHGEGDTGDHWLVVC--SGDSWVRTNPVRLQHIDTDMYL 179
Query: 407 HSHDKKYSRIAGGQQEVCAVREKRADNVWLAAEGVYLPVTE 285
+ + R GQ EV + + W+AAEG+++ TE
Sbjct: 180 GVSGRTFGRPINGQMEVIGLPNPYSGTEWIAAEGLFIHPTE 220
>ref|NP_649527.1| CG11999-PA [Drosophila melanogaster] gi|7296765|gb|AAF52043.1|
CG11999-PA [Drosophila melanogaster]
gi|21429978|gb|AAM50667.1| GH21273p [Drosophila
melanogaster]
Length = 216
Score = 83.2 bits (204), Expect = 2e-15
Identities = 39/96 (40%), Positives = 56/96 (57%)
Frame = -1
Query: 584 LHSHLHASPISGNLEVSCFGGESESDTGDYWKVFIEGSGKTWKQDQKIRLQHIDTGGYLH 405
LHSH +SP+SG EVS +G + DTGD+W+V S + W + +RL+HIDTG YL
Sbjct: 106 LHSHHFSSPLSGEQEVSAYGTDGLGDTGDHWEVVC--SNENWMRSAHVRLRHIDTGMYLG 163
Query: 404 SHDKKYSRIAGGQQEVCAVREKRADNVWLAAEGVYL 297
+ Y R GQ E+ V + + W AEG+++
Sbjct: 164 MSGRSYGRPISGQMEIVGVHKPQHGTRWTTAEGLFI 199
Score = 34.7 bits (78), Expect = 0.92
Identities = 26/83 (31%), Positives = 38/83 (45%), Gaps = 5/83 (6%)
Frame = -1
Query: 584 LHSHLHASPISGNLEVSCFGGESESDTGDYWKVFIEGS-----GKTWKQDQKIRLQHIDT 420
LHSH SG+ + S G E + D +W + + G+ +RL+H+ T
Sbjct: 44 LHSH-DVKYGSGSGQQSVTGVEQKEDVNSHWVIKAQTGELCERGEPIACGSTVRLEHLST 102
Query: 419 GGYLHSHDKKYSRIAGGQQEVCA 351
LHSH +S G+QEV A
Sbjct: 103 KKNLHSH--HFSSPLSGEQEVSA 123
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 516,684,990
Number of Sequences: 1393205
Number of extensions: 11149734
Number of successful extensions: 28700
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 27828
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 28682
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 22283372436
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)