Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC018169A_C01 KMC018169A_c01
(804 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
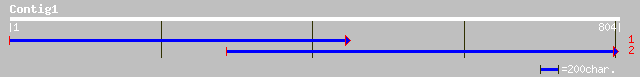
gb|AAN64490.1| unknown protein [Oryza sativa (japonica cultivar-... 44 0.003
ref|NP_195897.1| putative protein; protein id: At5g02770.1, supp... 43 0.006
gb|AAL10167.1| NADH dehydrogenase subunit 1 [Heloderma horridum] 34 2.9
gb|AAL10170.1| NADH dehydrogenase subunit 1 [Heloderma suspectum] 33 4.9
>gb|AAN64490.1| unknown protein [Oryza sativa (japonica cultivar-group)]
Length = 177
Score = 43.9 bits (102), Expect = 0.003
Identities = 27/55 (49%), Positives = 34/55 (61%), Gaps = 3/55 (5%)
Frame = -3
Query: 802 LARFAPPSKVDPLEEDKRKARALRFSNASSTPIPQVNGEGNVEP---KAAIAGNA 647
L RF + VD EE+KRKARALRF+ SS P Q NG+ + +P A +AG A
Sbjct: 124 LERFGQSTNVDKGEEEKRKARALRFAETSSGP-SQENGKDSSKPTQDAATVAGTA 177
>ref|NP_195897.1| putative protein; protein id: At5g02770.1, supported by cDNA:
gi_14190430 [Arabidopsis thaliana]
gi|11357918|pir||T48298 hypothetical protein F9G14.80 -
Arabidopsis thaliana gi|7413552|emb|CAB86031.1| putative
protein [Arabidopsis thaliana]
gi|14190431|gb|AAK55696.1|AF378893_1 AT5g02770/F9G14_80
[Arabidopsis thaliana] gi|15215889|gb|AAK91488.1|
AT5g02770/F9G14_80 [Arabidopsis thaliana]
Length = 214
Score = 42.7 bits (99), Expect = 0.006
Identities = 27/58 (46%), Positives = 36/58 (61%), Gaps = 6/58 (10%)
Frame = -3
Query: 802 LARFAPPSKVDPLEEDKRKARALRFSN------ASSTPIPQVNGEGNVEPKAAIAGNA 647
LARF +KVD EE+KRKARALRFS +S P Q+ G+ +AA++G+A
Sbjct: 161 LARFGKETKVDSAEENKRKARALRFSKEASADASSDLPEKQIIGK-----EAAVSGSA 213
>gb|AAL10167.1| NADH dehydrogenase subunit 1 [Heloderma horridum]
Length = 322
Score = 33.9 bits (76), Expect = 2.9
Identities = 17/38 (44%), Positives = 22/38 (57%)
Frame = -1
Query: 705 YLKSMERGMLNLRQLLRAMLEEGPESLLDPVAVGIFLF 592
YL +ER ML QL + GP+ LL P+A G+ LF
Sbjct: 24 YLTLLERKMLGYMQLRKGPNTVGPQGLLQPIADGVKLF 61
>gb|AAL10170.1| NADH dehydrogenase subunit 1 [Heloderma suspectum]
Length = 322
Score = 33.1 bits (74), Expect = 4.9
Identities = 17/38 (44%), Positives = 22/38 (57%)
Frame = -1
Query: 705 YLKSMERGMLNLRQLLRAMLEEGPESLLDPVAVGIFLF 592
YL +ER ML QL + GP+ LL P+A G+ LF
Sbjct: 24 YLTLLERKMLGYMQLRKGPNIVGPQGLLQPIADGVKLF 61
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 677,817,503
Number of Sequences: 1393205
Number of extensions: 14442822
Number of successful extensions: 36867
Number of sequences better than 10.0: 8
Number of HSP's better than 10.0 without gapping: 35235
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 36795
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 40896270532
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)