Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC017963A_C01 KMC017963A_c01
(533 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
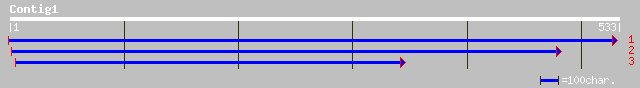
emb|CAC80857.1| C-type MADS box protein [Malus x domestica] 103 1e-21
dbj|BAA90743.1| MADS-box protein [Rosa rugosa] 103 2e-21
gb|AAK58564.1|AF265562_1 MAD-box transcripion factor [Vitis vini... 100 8e-21
gb|AAD38119.1| AGAMOUS homolog [Liquidambar styraciflua] 96 2e-19
gb|AAO18229.1| MADS-box transcriptional factor HAM59 [Helianthus... 92 3e-18
>emb|CAC80857.1| C-type MADS box protein [Malus x domestica]
Length = 242
Score = 103 bits (257), Expect = 1e-21
Identities = 53/80 (66%), Positives = 65/80 (81%), Gaps = 2/80 (2%)
Frame = -2
Query: 532 ESRLEKGLSRVRSRKHETLFGDIEFMQKREIELQNHNNLLRAKIAQHERAQQQEQS-LLQ 356
E RLEKG+SR+RS+K+E LF +IEFMQKRE ELQ+HNN LRAKIA+ ER QQQ+Q+ ++
Sbjct: 143 EGRLEKGISRIRSKKNEILFSEIEFMQKRETELQHHNNFLRAKIAESEREQQQQQTHMIP 202
Query: 355 GNPC-VSIPSQSYDRNFFPV 299
G S+PS SYDRNFFPV
Sbjct: 203 GTSYDPSMPSNSYDRNFFPV 222
>dbj|BAA90743.1| MADS-box protein [Rosa rugosa]
Length = 249
Score = 103 bits (256), Expect = 2e-21
Identities = 53/99 (53%), Positives = 73/99 (73%), Gaps = 4/99 (4%)
Frame = -2
Query: 532 ESRLEKGLSRVRSRKHETLFGDIEFMQKREIELQNHNNLLRAKIAQHERAQQQEQSLLQG 353
E RLEKG+SR+RS+K+E LF +IE+MQKREIELQNHNN LRAKIA+++RAQQQ+ +++ G
Sbjct: 147 EGRLEKGISRIRSKKNEMLFAEIEYMQKREIELQNHNNFLRAKIAENDRAQQQQANMMPG 206
Query: 352 NPCV----SIPSQSYDRNFFPVNVPNH*YSSCQDQICSP 248
P QSYDR+F PV + ++ + + Q Q +P
Sbjct: 207 TLSAYDQSMPPPQSYDRSFLPVILESNHHYNRQGQNQTP 245
>gb|AAK58564.1|AF265562_1 MAD-box transcripion factor [Vitis vinifera]
Length = 225
Score = 100 bits (250), Expect = 8e-21
Identities = 54/89 (60%), Positives = 70/89 (77%), Gaps = 3/89 (3%)
Frame = -2
Query: 532 ESRLEKGLSRVRSRKHETLFGDIEFMQKREIELQNHNNLLRAKIAQHERAQQQEQSLLQG 353
E+RLEKG+SR+RS+K+E LF +IE+MQKREIELQN N LRA+IA++ERAQQQ +L+ G
Sbjct: 128 ETRLEKGISRIRSKKNELLFAEIEYMQKREIELQNSNLFLRAQIAENERAQQQ-MNLMPG 186
Query: 352 NPCVSIPSQSYD-RNFFPVNV--PNH*YS 275
+ S+P Q YD +N PVN+ PNH YS
Sbjct: 187 SQYESVPQQPYDSQNLLPVNLLDPNHHYS 215
>gb|AAD38119.1| AGAMOUS homolog [Liquidambar styraciflua]
Length = 244
Score = 96.3 bits (238), Expect = 2e-19
Identities = 56/95 (58%), Positives = 71/95 (73%), Gaps = 4/95 (4%)
Frame = -2
Query: 532 ESRLEKGLSRVRSRKHETLFGDIEFMQKREIELQNHNNLLRAKIAQHERAQQQEQSLLQG 353
E RLEKG+SR+RS+K+E LF +IE+MQKREIELQN N LRAKIA++ER QQQ + L+ G
Sbjct: 146 EGRLEKGISRIRSKKNELLFAEIEYMQKREIELQNANMYLRAKIAENERNQQQTE-LMPG 204
Query: 352 NPCVSIP-SQSYDRNFFPVNV---PNH*YSSCQDQ 260
+ ++P SQ YDR+F N+ PNH YS QDQ
Sbjct: 205 SVYETMPSSQPYDRSFLVANLLEPPNHHYSR-QDQ 238
>gb|AAO18229.1| MADS-box transcriptional factor HAM59 [Helianthus annuus]
Length = 247
Score = 92.4 bits (228), Expect = 3e-18
Identities = 52/96 (54%), Positives = 69/96 (71%), Gaps = 5/96 (5%)
Frame = -2
Query: 532 ESRLEKGLSRVRSRKHETLFGDIEFMQKREIELQNHNNLLRAKIAQHERAQQQEQSLLQG 353
E +LEKG+SR+RS+K+E LF +IE+M KRE EL N+N LRAKIA++ER+QQQ SL+ G
Sbjct: 147 EGKLEKGISRIRSKKNELLFAEIEYMPKRENELHNNNQFLRAKIAENERSQQQHMSLMPG 206
Query: 352 NPCVSI--PSQSYD-RNFFPVN--VPNH*YSSCQDQ 260
+ + P Q +D RN+ VN PN+ Y SCQDQ
Sbjct: 207 SSDYDLVPPHQPFDGRNYLQVNDLQPNNSY-SCQDQ 241
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 438,306,277
Number of Sequences: 1393205
Number of extensions: 8993453
Number of successful extensions: 25807
Number of sequences better than 10.0: 227
Number of HSP's better than 10.0 without gapping: 24525
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25720
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 17885181664
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)