Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC017790A_C01 KMC017790A_c01
(1020 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
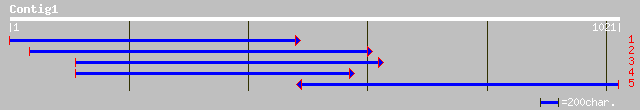
gb|AAG24946.1| dehydroascorbate reductase [Arabidopsis thaliana]... 354 1e-96
ref|NP_568336.1| dehydroascorbate reductase, putative; protein i... 354 1e-96
gb|AAN04049.1| dehydroascorbate reductase [Brassica juncea] 353 2e-96
gb|AAL38300.1| unknown protein [Arabidopsis thaliana] gi|2489971... 351 1e-95
gb|AAN04048.1| dehydroascorbate reductase [Brassica juncea] 350 3e-95
>gb|AAG24946.1| dehydroascorbate reductase [Arabidopsis thaliana]
gi|11761812|gb|AAG40196.1|AF301597_1 glutathione
dependent dehydroascorbate reductase precursor
[Arabidopsis thaliana]
Length = 252
Score = 354 bits (909), Expect = 1e-96
Identities = 165/229 (72%), Positives = 195/229 (85%), Gaps = 2/229 (0%)
Frame = -1
Query: 972 SRKPLRVS--MSSVPPSEPVEIAVKASLTTPNTLGDCPFSQRVLLTLEEKHLPYEAKFVD 799
S KP RV ++ + P+EI VKAS+TTPN LGDCPF Q+VLLT+EEK++PY+ K VD
Sbjct: 24 STKPGRVGRFVTMATAASPLEICVKASITTPNKLGDCPFCQKVLLTMEEKNVPYDMKMVD 83
Query: 798 LSNKPEWFLQISPEGKVPVIKFDEKWVPDSDIITQTXEEKYPSPPXVTPPEKSTVGSKIF 619
LSNKPEWFL+ISPEGKVPV+KFDEKWVPDSD+ITQ EEKYP PP TPPEK++VGSKIF
Sbjct: 84 LSNKPEWFLKISPEGKVPVVKFDEKWVPDSDVITQALEEKYPEPPLATPPEKASVGSKIF 143
Query: 618 STFIGFLKSKDPNDGTEQALLSELSSFNDYLKDNGPFVNGKDISAADLSLGPKLYHLEIA 439
STF+GFLKSKD DGTEQ LL EL++FNDY+KDNGPF+NG+ ISAADLSL PKLYH++IA
Sbjct: 144 STFVGFLKSKDSGDGTEQVLLDELTTFNDYIKDNGPFINGEKISAADLSLAPKLYHMKIA 203
Query: 438 LGHYKKWTVPDSLTFLKSYMKAIFSRESFIKTRAQPQDVVEGWRPKVEG 292
LGHYK W+VPDSL F+KSYM+ +FSRESF TRA+ +DV+ GWRPKV G
Sbjct: 204 LGHYKNWSVPDSLPFVKSYMENVFSRESFTNTRAETEDVIAGWRPKVMG 252
>ref|NP_568336.1| dehydroascorbate reductase, putative; protein id: At5g16710.1,
supported by cDNA: 16205., supported by cDNA:
gi_17473686 [Arabidopsis thaliana]
gi|21553744|gb|AAM62837.1| dehydroascorbate reductase
[Arabidopsis thaliana] gi|26452229|dbj|BAC43202.1|
putative dehydroascorbate reductase [Arabidopsis
thaliana]
Length = 258
Score = 354 bits (909), Expect = 1e-96
Identities = 165/229 (72%), Positives = 195/229 (85%), Gaps = 2/229 (0%)
Frame = -1
Query: 972 SRKPLRVS--MSSVPPSEPVEIAVKASLTTPNTLGDCPFSQRVLLTLEEKHLPYEAKFVD 799
S KP RV ++ + P+EI VKAS+TTPN LGDCPF Q+VLLT+EEK++PY+ K VD
Sbjct: 30 STKPGRVGRFVTMATAASPLEICVKASITTPNKLGDCPFCQKVLLTMEEKNVPYDMKMVD 89
Query: 798 LSNKPEWFLQISPEGKVPVIKFDEKWVPDSDIITQTXEEKYPSPPXVTPPEKSTVGSKIF 619
LSNKPEWFL+ISPEGKVPV+KFDEKWVPDSD+ITQ EEKYP PP TPPEK++VGSKIF
Sbjct: 90 LSNKPEWFLKISPEGKVPVVKFDEKWVPDSDVITQALEEKYPEPPLATPPEKASVGSKIF 149
Query: 618 STFIGFLKSKDPNDGTEQALLSELSSFNDYLKDNGPFVNGKDISAADLSLGPKLYHLEIA 439
STF+GFLKSKD DGTEQ LL EL++FNDY+KDNGPF+NG+ ISAADLSL PKLYH++IA
Sbjct: 150 STFVGFLKSKDSGDGTEQVLLDELTTFNDYIKDNGPFINGEKISAADLSLAPKLYHMKIA 209
Query: 438 LGHYKKWTVPDSLTFLKSYMKAIFSRESFIKTRAQPQDVVEGWRPKVEG 292
LGHYK W+VPDSL F+KSYM+ +FSRESF TRA+ +DV+ GWRPKV G
Sbjct: 210 LGHYKNWSVPDSLPFVKSYMENVFSRESFTNTRAETEDVIAGWRPKVMG 258
>gb|AAN04049.1| dehydroascorbate reductase [Brassica juncea]
Length = 217
Score = 353 bits (906), Expect = 2e-96
Identities = 163/213 (76%), Positives = 189/213 (88%)
Frame = -1
Query: 930 SEPVEIAVKASLTTPNTLGDCPFSQRVLLTLEEKHLPYEAKFVDLSNKPEWFLQISPEGK 751
S P+EI VKAS+TTPN LGDCPF QRVLLT+EEKH+PY+ K VDLSNKPEWFL+IS EGK
Sbjct: 5 SAPLEICVKASITTPNKLGDCPFCQRVLLTMEEKHVPYDMKMVDLSNKPEWFLKISAEGK 64
Query: 750 VPVIKFDEKWVPDSDIITQTXEEKYPSPPXVTPPEKSTVGSKIFSTFIGFLKSKDPNDGT 571
VPV+KFDEKWVPDSD+ITQ+ E+KYP PP TPPEK++VGSKIFSTFIGFLKSKD DGT
Sbjct: 65 VPVVKFDEKWVPDSDVITQSLEDKYPEPPLATPPEKASVGSKIFSTFIGFLKSKDSGDGT 124
Query: 570 EQALLSELSSFNDYLKDNGPFVNGKDISAADLSLGPKLYHLEIALGHYKKWTVPDSLTFL 391
EQ LL ELS+FNDYLK+NGP++NG+ ISAADLSL PKLYH++IALGH+K W+VPDSL+FL
Sbjct: 125 EQVLLDELSTFNDYLKENGPYINGEKISAADLSLAPKLYHMKIALGHFKNWSVPDSLSFL 184
Query: 390 KSYMKAIFSRESFIKTRAQPQDVVEGWRPKVEG 292
KSYM+ +FSRESF KT AQ +DV+ GWRPKV G
Sbjct: 185 KSYMENVFSRESFKKTEAQTEDVIAGWRPKVMG 217
>gb|AAL38300.1| unknown protein [Arabidopsis thaliana] gi|24899715|gb|AAN65072.1|
unknown protein [Arabidopsis thaliana]
Length = 258
Score = 351 bits (900), Expect = 1e-95
Identities = 164/229 (71%), Positives = 194/229 (84%), Gaps = 2/229 (0%)
Frame = -1
Query: 972 SRKPLRVS--MSSVPPSEPVEIAVKASLTTPNTLGDCPFSQRVLLTLEEKHLPYEAKFVD 799
S KP RV ++ + P+EI VKAS+TTPN LG CPF Q+VLLT+EEK++PY+ K VD
Sbjct: 30 STKPGRVGRFVTMATAASPLEICVKASITTPNKLGYCPFCQKVLLTMEEKNVPYDMKMVD 89
Query: 798 LSNKPEWFLQISPEGKVPVIKFDEKWVPDSDIITQTXEEKYPSPPXVTPPEKSTVGSKIF 619
LSNKPEWFL+ISPEGKVPV+KFDEKWVPDSD+ITQ EEKYP PP TPPEK++VGSKIF
Sbjct: 90 LSNKPEWFLKISPEGKVPVVKFDEKWVPDSDVITQALEEKYPEPPLATPPEKASVGSKIF 149
Query: 618 STFIGFLKSKDPNDGTEQALLSELSSFNDYLKDNGPFVNGKDISAADLSLGPKLYHLEIA 439
STF+GFLKSKD DGTEQ LL EL++FNDY+KDNGPF+NG+ ISAADLSL PKLYH++IA
Sbjct: 150 STFVGFLKSKDSGDGTEQVLLDELTTFNDYIKDNGPFINGEKISAADLSLAPKLYHMKIA 209
Query: 438 LGHYKKWTVPDSLTFLKSYMKAIFSRESFIKTRAQPQDVVEGWRPKVEG 292
LGHYK W+VPDSL F+KSYM+ +FSRESF TRA+ +DV+ GWRPKV G
Sbjct: 210 LGHYKNWSVPDSLPFVKSYMENVFSRESFTNTRAETEDVIAGWRPKVMG 258
>gb|AAN04048.1| dehydroascorbate reductase [Brassica juncea]
Length = 257
Score = 350 bits (897), Expect = 3e-95
Identities = 163/229 (71%), Positives = 194/229 (84%)
Frame = -1
Query: 978 THSRKPLRVSMSSVPPSEPVEIAVKASLTTPNTLGDCPFSQRVLLTLEEKHLPYEAKFVD 799
T R+ +R+ ++ + P+EI VKAS+TTPN LGDCPF QRVLLT+EEKH+PY+ K VD
Sbjct: 30 TKPRRTVRLGTVAMAAA-PLEICVKASITTPNKLGDCPFCQRVLLTMEEKHVPYDMKMVD 88
Query: 798 LSNKPEWFLQISPEGKVPVIKFDEKWVPDSDIITQTXEEKYPSPPXVTPPEKSTVGSKIF 619
LSNKPEWFL+I+ EGKVPV+KFDEKWVPDSD+IT E+KYP PP TPPEK++VGSKIF
Sbjct: 89 LSNKPEWFLKINAEGKVPVVKFDEKWVPDSDVITHALEDKYPEPPLATPPEKASVGSKIF 148
Query: 618 STFIGFLKSKDPNDGTEQALLSELSSFNDYLKDNGPFVNGKDISAADLSLGPKLYHLEIA 439
STFIGFLKSKDP DGTEQ LL ELS+FNDYLK+NGP++NG+ ISAADLSL PKLYH++IA
Sbjct: 149 STFIGFLKSKDPKDGTEQVLLDELSTFNDYLKENGPYINGEKISAADLSLAPKLYHMKIA 208
Query: 438 LGHYKKWTVPDSLTFLKSYMKAIFSRESFIKTRAQPQDVVEGWRPKVEG 292
LGH+K W+VPDSL FLKSYM+ +FSRESF T AQ +DV+ GWRPKV G
Sbjct: 209 LGHFKNWSVPDSLPFLKSYMENVFSRESFKNTEAQTEDVIAGWRPKVMG 257
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 942,918,519
Number of Sequences: 1393205
Number of extensions: 21874896
Number of successful extensions: 61612
Number of sequences better than 10.0: 416
Number of HSP's better than 10.0 without gapping: 56442
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 61143
length of database: 448,689,247
effective HSP length: 124
effective length of database: 275,931,827
effective search space used: 59325342805
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)