Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC017626A_C01 KMC017626A_c01
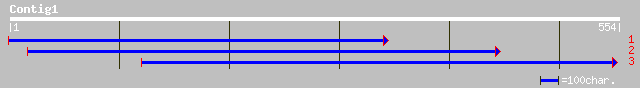
(554 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_295043.1| hypothetical protein [Deinococcus radiodurans] ... 33 1.8
ref|XP_223489.1| similar to high mobility group protein 14 [Mus ... 33 2.3
ref|NP_659289.1| NADH dehydrogenase subunit 2 [Lemur catta] gi|2... 32 6.8
ref|NP_066452.1| orf154 [Ochromonas danica] gi|11465906|ref|NP_0... 32 6.8
emb|CAD60557.1| unnamed protein product [Podospora anserina] 31 8.9
>ref|NP_295043.1| hypothetical protein [Deinococcus radiodurans]
gi|7472323|pir||C75411 hypothetical protein -
Deinococcus radiodurans (strain R1)
gi|6459072|gb|AAF10893.1|AE001978_13 hypothetical
protein [Deinococcus radiodurans]
Length = 200
Score = 33.5 bits (75), Expect = 1.8
Identities = 17/59 (28%), Positives = 35/59 (58%), Gaps = 2/59 (3%)
Frame = -2
Query: 547 DGRITNNGKELKRGIKARRKEREKRVFFN--CKIIQESNA*RLSINEAESIMQHLMTLV 377
D ++ +G RG+K+R + E RV+F+ ++I++ N ++ EA +++Q TL+
Sbjct: 131 DIQVNKDGSITDRGVKSRAQTAEMRVYFDDQGRVIRQLNKFPMTKQEAGALLQEFRTLI 189
>ref|XP_223489.1| similar to high mobility group protein 14 [Mus musculus] [Rattus
norvegicus]
Length = 110
Score = 33.1 bits (74), Expect = 2.3
Identities = 22/71 (30%), Positives = 35/71 (48%), Gaps = 6/71 (8%)
Frame = -1
Query: 512 ERHKSKEKRKGEKSFLQLQDYSGIKCLEIVNK*GRINHAALDDLGSIYSCEKRIQ----- 348
ER K + K++G Q++ +K LE K ++ H DLG++Y C+K +
Sbjct: 41 ERLKHQTKKQGRSKHKQIR----VKALEQFTKVDKLYHNRFYDLGALYFCQKVSEKAEAG 96
Query: 347 -GGENEAAEAT 318
G+ AAE T
Sbjct: 97 DAGKRMAAEET 107
>ref|NP_659289.1| NADH dehydrogenase subunit 2 [Lemur catta]
gi|21425355|emb|CAD13422.1| NADH dehydrogenase subunit 2
[Lemur catta]
Length = 347
Score = 31.6 bits (70), Expect = 6.8
Identities = 14/45 (31%), Positives = 26/45 (57%)
Frame = -2
Query: 160 LFTFIIFFPSFSVINQFIFLFLLKYVFSVAQINSLKKTRSNQ*TW 26
+ ++++PS +++N I+L L +FSV IN+ T + TW
Sbjct: 190 MMAILMYWPSLTMLNLLIYLMLTITLFSVLNINTNTTTLTLSNTW 234
>ref|NP_066452.1| orf154 [Ochromonas danica] gi|11465906|ref|NP_066411.1| orf154
[Ochromonas danica]
gi|10505236|gb|AAG18418.1|AF287134_43 orf154 [Ochromonas
danica] gi|10505239|gb|AAG18421.1| orf154 [Ochromonas
danica]
Length = 154
Score = 31.6 bits (70), Expect = 6.8
Identities = 18/49 (36%), Positives = 31/49 (62%), Gaps = 5/49 (10%)
Frame = -2
Query: 181 FCSIYL*LFTFII-----FFPSFSVINQFIFLFLLKYVFSVAQINSLKK 50
FC++Y+ LF++ I + P + +I +F+FLL Y FS+ NS++K
Sbjct: 97 FCNMYI-LFSYSIDELPFWLPIYYIILSMLFVFLLGYTFSI-HSNSIEK 143
>emb|CAD60557.1| unnamed protein product [Podospora anserina]
Length = 668
Score = 31.2 bits (69), Expect = 8.9
Identities = 29/90 (32%), Positives = 41/90 (45%), Gaps = 5/90 (5%)
Frame = +1
Query: 160 VRDKWSKRINKDRSSLDFMLQATFLGKYKSANEWMIPLEIFFRSSTF--YLIFNEVASA- 330
V+ WS I S F+L A F+ K E ++PLEIF + YLI +A
Sbjct: 387 VKLPWSSPITISLISFSFVLIAVFVAVEKRQEEPILPLEIFHQRDAVISYLILGLQTAAQ 446
Query: 331 --ASFSPPCILFSQL*IEPRSSSAA*LILP 414
FS P L+ Q+ +SSA ++P
Sbjct: 447 LGLMFSVP--LYFQITSRSSASSAGAHLVP 474
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 412,438,102
Number of Sequences: 1393205
Number of extensions: 7790789
Number of successful extensions: 22308
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 21631
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 22287
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19521267756
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)