Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC017314A_C01 KMC017314A_c01
(589 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
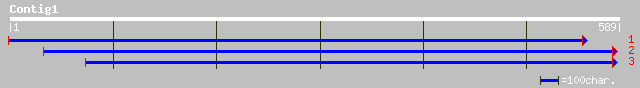
ref|NP_200665.2| putative protein; protein id: At5g58575.1, supp... 90 2e-17
dbj|BAA97326.1| emb|CAB85555.1~gene_id:MZN1.2~similar to unknown... 90 2e-17
ref|NP_649050.1| CG13379-PA [Drosophila melanogaster] gi|7293878... 40 0.022
emb|CAD43430.2| SI:dZ180G5.4 (novel protein) [Danio rerio] 39 0.037
gb|AAH37418.1| Unknown (protein for IMAGE:5499446) [Homo sapiens] 39 0.063
>ref|NP_200665.2| putative protein; protein id: At5g58575.1, supported by cDNA:
gi_14532481, supported by cDNA: gi_18655362 [Arabidopsis
thaliana] gi|14532482|gb|AAK63969.1| AT5g58570/mzn1_20
[Arabidopsis thaliana] gi|18655363|gb|AAL76137.1|
AT5g58570/mzn1_20 [Arabidopsis thaliana]
Length = 181
Score = 89.7 bits (221), Expect = 2e-17
Identities = 48/77 (62%), Positives = 54/77 (69%)
Frame = -3
Query: 587 GRSIMAGRFAPHLEKCMGKGRKARLKVTRSSTATQKRYSRGIPASRYSSYSNHSSNSINR 408
GR I+AGRFAPHLEKCMGKGRKAR K TRS+TA Q R +R P RYS Y N +S N+
Sbjct: 99 GRQIVAGRFAPHLEKCMGKGRKARAKTTRSTTAAQNRNARRSPNPRYSPYPNSASE--NQ 156
Query: 407 SANGSSSFAGEGHSNGT 357
A+GS AGE SN T
Sbjct: 157 LASGSPGVAGEDCSNFT 173
>dbj|BAA97326.1| emb|CAB85555.1~gene_id:MZN1.2~similar to unknown protein
[Arabidopsis thaliana]
Length = 517
Score = 89.7 bits (221), Expect = 2e-17
Identities = 48/77 (62%), Positives = 54/77 (69%)
Frame = -3
Query: 587 GRSIMAGRFAPHLEKCMGKGRKARLKVTRSSTATQKRYSRGIPASRYSSYSNHSSNSINR 408
GR I+AGRFAPHLEKCMGKGRKAR K TRS+TA Q R +R P RYS Y N +S N+
Sbjct: 435 GRQIVAGRFAPHLEKCMGKGRKARAKTTRSTTAAQNRNARRSPNPRYSPYPNSASE--NQ 492
Query: 407 SANGSSSFAGEGHSNGT 357
A+GS AGE SN T
Sbjct: 493 LASGSPGVAGEDCSNFT 509
>ref|NP_649050.1| CG13379-PA [Drosophila melanogaster] gi|7293878|gb|AAF49243.1|
CG13379-PA [Drosophila melanogaster]
Length = 223
Score = 40.0 bits (92), Expect = 0.022
Identities = 26/81 (32%), Positives = 36/81 (44%), Gaps = 6/81 (7%)
Frame = -3
Query: 584 RSIMAGRFAPHLEKCMGKGR------KARLKVTRSSTATQKRYSRGIPASRYSSYSNHSS 423
R + A RFAPHLEKCMG GR RL +T+ S + + SS
Sbjct: 113 RLVAAARFAPHLEKCMGMGRISSRIASRRLATKEGATSAHLHSSGNTGGTDDEDDVDWSS 172
Query: 422 NSINRSANGSSSFAGEGHSNG 360
+ + +N +S G +NG
Sbjct: 173 DKRRKKSNQNSRNNGSKKNNG 193
>emb|CAD43430.2| SI:dZ180G5.4 (novel protein) [Danio rerio]
Length = 335
Score = 39.3 bits (90), Expect = 0.037
Identities = 22/41 (53%), Positives = 24/41 (57%), Gaps = 3/41 (7%)
Frame = -3
Query: 584 RSIMAGRFAPHLEKCMGKGRKARLKVTR---SSTATQKRYS 471
RSI A RFAPHLEKC+G GR + R SS T K S
Sbjct: 91 RSIAASRFAPHLEKCLGMGRNSSRIANRRIASSNNTSKSES 131
>gb|AAH37418.1| Unknown (protein for IMAGE:5499446) [Homo sapiens]
Length = 353
Score = 38.5 bits (88), Expect = 0.063
Identities = 16/22 (72%), Positives = 18/22 (81%)
Frame = -3
Query: 584 RSIMAGRFAPHLEKCMGKGRKA 519
RSI A RFAPHLEKC+G GR +
Sbjct: 90 RSIAASRFAPHLEKCLGMGRNS 111
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 500,663,861
Number of Sequences: 1393205
Number of extensions: 10388992
Number of successful extensions: 29437
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 27323
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 29264
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 22283372436
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)