Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC017298A_C01 KMC017298A_c01
(1469 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
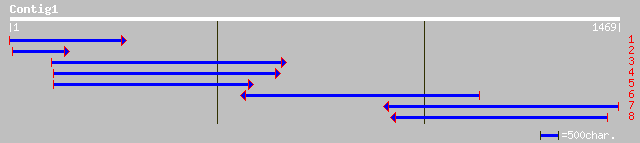
ref|NP_200423.1| putative protein; protein id: At5g56120.1 [Arab... 277 3e-73
dbj|BAA81760.1| hypothetical protein [Oryza sativa (japonica cul... 182 9e-45
ref|NP_187893.1| hypothetical protein; protein id: At3g12870.1 [... 134 5e-30
gb|AAK09219.1|AC084320_6 hypothetical protein [Oryza sativa (jap... 119 1e-25
dbj|BAA81756.1| hypothetical protein [Oryza sativa (japonica cul... 94 7e-18
>ref|NP_200423.1| putative protein; protein id: At5g56120.1 [Arabidopsis thaliana]
gi|9758632|dbj|BAB09294.1|
dbj|BAA81760.1~gene_id:MDA7.18~similar to unknown protein
[Arabidopsis thaliana]
Length = 237
Score = 277 bits (709), Expect = 3e-73
Identities = 133/212 (62%), Positives = 165/212 (77%), Gaps = 7/212 (3%)
Frame = -2
Query: 1366 MAETRDTHIVEIPVDQEHKHH-----LCSMTTTNM--FEAIDDHPLTEISESPGHLLLLK 1208
MA T+D+H+VEIPVD+EHK L +++ M + I HPL+EISESPGHLLLLK
Sbjct: 1 MAGTKDSHVVEIPVDEEHKERQQQQQLVAVSNPGMGILKVIQQHPLSEISESPGHLLLLK 60
Query: 1207 LWQREEDLFAKRIARKETRMDSVKAELFQLCSFFFIFHGFFFTLLFTSWTSEQNHPDQHH 1028
LWQREEDLF +R+ KE+R++S+K E+FQLC FF +FHGFFFTL+++S S+ + +
Sbjct: 61 LWQREEDLFCRRVLLKESRLESIKREIFQLCCFFLVFHGFFFTLVYSSSCSDDADVVKSN 120
Query: 1027 SVCKKWWIPSMVSLCTSLVFVFLVQVKVHGYWKVWEQLQRERNDGRAVTRCIQELRMKGA 848
+VCKKWWIPS VSL TSLV VFLVQ K+ +WKV+ + RERND R +TRC+ ELRMKG+
Sbjct: 121 AVCKKWWIPSAVSLATSLVLVFLVQAKLFVFWKVYRGVNRERNDNRTLTRCVLELRMKGS 180
Query: 847 SFDLSKEPQSGKRMKSSSVEIKWKPVTWCSHN 752
SFDLSKEP SGKRMKSSSVEIKWKPVTW H+
Sbjct: 181 SFDLSKEPMSGKRMKSSSVEIKWKPVTWTLHS 212
>dbj|BAA81760.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 380
Score = 182 bits (463), Expect = 9e-45
Identities = 100/233 (42%), Positives = 129/233 (54%), Gaps = 13/233 (5%)
Frame = -2
Query: 1345 HIVEIPV-------DQEHKHHLCSMTTTNMFEAIDDHPLTEISESPGHLLLLKLWQREED 1187
HIVEIPV D + + E+ HPL EI+ S GHLLLLKLWQREE
Sbjct: 147 HIVEIPVTGDGACPDGGNAEVAAFLDKAAEAESSGSHPLGEIAGSAGHLLLLKLWQREES 206
Query: 1186 LFAKRIARKETRMDSVKAELFQLCSFFFIFHGFFFTLLFTSWTSEQN---HPDQHHSVCK 1016
+R E MD+ + + F LC+ F FHG LLF + S +Q + C
Sbjct: 207 RLGRRACALEALMDAARRDAFYLCAAFLAFHGLSLALLFAASVSASAVSPPAEQRAACCS 266
Query: 1015 KWWIPSMVSLCTSLVFVFLVQVKVHGYWKVWEQLQRERNDGRAVTRCIQELRMKGASFDL 836
+WW+PS +SL SL VQ++V YW+ +L+RER D RA+ RC+QELRMKGA+FDL
Sbjct: 267 RWWVPSSLSLVASLALAAAVQLRVCAYWRASRRLRRERGDARALARCVQELRMKGAAFDL 326
Query: 835 SKEPQSG-KRMKSSSVE--IKWKPVTWCSHNLLTFCLVCFTGLAFPVSKFVLC 686
SKEPQ G R K +SVE W P+ WC N++ CL+ KF+LC
Sbjct: 327 SKEPQYGVTRAKCASVEGAGAWGPLRWCYQNIVAACLLAVAAATMCSGKFILC 379
>ref|NP_187893.1| hypothetical protein; protein id: At3g12870.1 [Arabidopsis thaliana]
gi|6997162|gb|AAF34826.1| hypothetical protein
[Arabidopsis thaliana] gi|9279771|dbj|BAB01416.1|
gb|AAF34826.1~gene_id:MJM20.1~similar to unknown protein
[Arabidopsis thaliana]
Length = 206
Score = 134 bits (336), Expect = 5e-30
Identities = 74/203 (36%), Positives = 118/203 (57%), Gaps = 1/203 (0%)
Frame = -2
Query: 1291 TTTNMFEAIDDHPLTEISESPGHLLLLKLWQREEDLFAKRIARKETRMDSVKAELFQLCS 1112
T + EA HPL +I+++P H LLLK W +EE+L R++ KE+++DSV+ E+ QL
Sbjct: 7 TNPPIMEAKTRHPLHQIADTPTHKLLLKQWLKEEELILSRVSHKESQIDSVRREITQLYI 66
Query: 1111 FFFIFHGFFFTLLFTSWTSEQNHPDQHHSVCKKWWIPSMVSLCTSLVFVFLVQVKVHGYW 932
FFF+FH LLF + +S + S CK+ WIPS+ +L +SL ++ V+ K
Sbjct: 67 FFFLFHSISLLLLFHASSSSSSSASS--SACKRSWIPSLCALLSSLGIIWAVRYKSEVES 124
Query: 931 KVWEQLQRERNDGRAVTRCIQELRMKGASFDLSKEPQSGKRMKSSSVEIKWKPV-TWCSH 755
+ + L+RE+ D + + +C++EL+ KG FDL KE + +R KS VE KPV W +
Sbjct: 125 HLEKLLEREKEDAKLLRKCVEELKKKGIEFDLLKEVDALRRAKSLRVES--KPVKKWSAR 182
Query: 754 NLLTFCLVCFTGLAFPVSKFVLC 686
+ +T + L + + +LC
Sbjct: 183 DFVTLFFFSVSCLVLAMIRLILC 205
>gb|AAK09219.1|AC084320_6 hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|20303603|gb|AAM19030.1|AC084748_20 hypothetical
protein [Oryza sativa (japonica cultivar-group)]
Length = 241
Score = 119 bits (298), Expect = 1e-25
Identities = 68/180 (37%), Positives = 100/180 (54%), Gaps = 5/180 (2%)
Frame = -2
Query: 1258 HPLTEISESPGHLLLLKLWQREEDLFAKRIARKETRMDSVKAELFQLCSFFFIFHGFFFT 1079
HPL++I+ES H LLLK W +EEDL A+R+A +E R+D + E+ L FF FH
Sbjct: 16 HPLSQIAESGTHRLLLKQWLKEEDLLARRVALREARLDGARKEIAFLYCAFFAFHAASIL 75
Query: 1078 LLFTSWTSEQNHPDQHHSVCKKWWIPSMVSLCTSLVFVFLVQVKVHGYWKVWEQLQRERN 899
LLF S ++ + + C++ WIP +VSL +SL ++ ++ K + L RER
Sbjct: 76 LLFLSASASTS------AACRRSWIPCLVSLLSSLAMLWALRYKADTEAVLERLLARERE 129
Query: 898 DGRAVTRCIQELRMKGASFDLSKEPQSGKRMKSSSVEIKW-----KPVTWCSHNLLTFCL 734
D + +C+ EL+ KG FDL KE + +R KS VE K +P W + +L F L
Sbjct: 130 DALLLGKCVAELKRKGLEFDLLKEVDALRRAKSLRVEAKGGAGGERPKRWAARDLAVFLL 189
>dbj|BAA81756.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 253
Score = 93.6 bits (231), Expect = 7e-18
Identities = 63/172 (36%), Positives = 79/172 (45%), Gaps = 19/172 (11%)
Frame = -2
Query: 1273 EAIDDHPLTEISESPGHLLLLKLWQREEDLFAKRIARKETRMDSVKAELFQLCSFFFIFH 1094
E+ HPL EI+ S GHLLLLKLWQREE +R E MD+ + + F LC+ F FH
Sbjct: 51 ESSGSHPLGEIAGSAGHLLLLKLWQREESRLGRRACALEALMDAARRDAFYLCAAFLAFH 110
Query: 1093 GFFFTLLFTSWTSEQ------NHPDQHHSVC------------KKWWIPSMVSL-CTSLV 971
G LLF + S + P + + C +W PS C+S
Sbjct: 111 GLSLALLFAASVSASASAAVVSPPAEQRAACCSLAGGGGCRRPCRWSRPSRSRRPCSS-- 168
Query: 970 FVFLVQVKVHGYWKVWEQLQRERNDGRAVTRCIQELRMKGASFDLSKEPQSG 815
R RA+ RC+QELRMKGA+F LSKEPQ G
Sbjct: 169 ----------------GSAPGVRTATRALARCVQELRMKGAAFGLSKEPQYG 204
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,337,835,766
Number of Sequences: 1393205
Number of extensions: 32530670
Number of successful extensions: 190328
Number of sequences better than 10.0: 334
Number of HSP's better than 10.0 without gapping: 115321
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 168729
length of database: 448,689,247
effective HSP length: 127
effective length of database: 271,752,212
effective search space used: 98374300744
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)