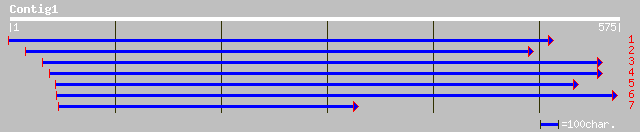
Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC017227A_C01 KMC017227A_c01
(575 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sp|P16059|PSBP_PEA Oxygen-evolving enhancer protein 2, chloropla... 153 1e-36
sp|P12302|PSBP_SPIOL Oxygen-evolving enhancer protein 2, chlorop... 152 3e-36
pir||T02873 probable photosystem II oxygen-evolving complex prot... 151 5e-36
sp|O49080|PSBP_FRIAG Oxygen-evolving enhancer protein 2, chlorop... 146 2e-34
sp|Q40407|PSBP_NARPS Oxygen-evolving enhancer protein 2, chlorop... 145 3e-34
>sp|P16059|PSBP_PEA Oxygen-evolving enhancer protein 2, chloroplast precursor (OEE2)
(23 kDa subunit of oxygen evolving system of photosystem
II) (OEC 23 kDa subunit) (23 kDa thylakoid membrane
protein) gi|280396|pir||JS0771 photosystem II
oxygen-evolving complex protein 2 precursor - garden pea
gi|20617|emb|CAA33557.1| precursor polypeptide (AA -73
to 186) [Pisum sativum] gi|344006|dbj|BAA02553.1|
precursor for 23-kDa protein of photosystem II [Pisum
sativum]
Length = 259
Score = 153 bits (387), Expect = 1e-36
Identities = 74/87 (85%), Positives = 79/87 (90%)
Frame = -2
Query: 574 EGGFDPNAVATANILESSTPVVDGKQYYVLTVLTRTADGDEGGKHQLIRAAVKDGKLYIC 395
EGGFD NAVA ANILESS PV+ GKQYY ++VLTRTADGDEGGKHQLI A VKDGKLYIC
Sbjct: 173 EGGFDTNAVAVANILESSAPVIGGKQYYNISVLTRTADGDEGGKHQLITATVKDGKLYIC 232
Query: 394 KAQAGDKRWFKGARRYVESTASSFSVA 314
KAQAGDKRWFKGAR++VE TASSFSVA
Sbjct: 233 KAQAGDKRWFKGARKFVEDTASSFSVA 259
>sp|P12302|PSBP_SPIOL Oxygen-evolving enhancer protein 2, chloroplast precursor (OEE2)
(23 kDa subunit of oxygen evolving system of photosystem
II) (OEC 23 kDa subunit) (23 kDa thylakoid membrane
protein) gi|81479|pir||S00005 photosystem II
oxygen-evolving complex protein 2 precursor - spinach
gi|21265|emb|CAA29055.1| 23 kDa OEC protein [Spinacia
oleracea] gi|225596|prf||1307179A luminal protein 23kD
Length = 267
Score = 152 bits (384), Expect = 3e-36
Identities = 72/87 (82%), Positives = 79/87 (90%)
Frame = -2
Query: 574 EGGFDPNAVATANILESSTPVVDGKQYYVLTVLTRTADGDEGGKHQLIRAAVKDGKLYIC 395
EGGFD VA+AN+LESSTPVVDGKQYY +TVLTRTADGDEGGKHQ+I A VKDGKLYIC
Sbjct: 181 EGGFDSGVVASANVLESSTPVVDGKQYYSITVLTRTADGDEGGKHQVIAATVKDGKLYIC 240
Query: 394 KAQAGDKRWFKGARRYVESTASSFSVA 314
KAQAGDKRWFKGA+++VES SSFSVA
Sbjct: 241 KAQAGDKRWFKGAKKFVESATSSFSVA 267
>pir||T02873 probable photosystem II oxygen-evolving complex protein 2 precursor
- rice gi|4079798|gb|AAC98778.1| 23 kDa polypeptide of
photosystem II [Oryza sativa]
gi|24059930|dbj|BAC21393.1| probable photosystem II
oxygen-evolving complex protein 2 precursor [Oryza
sativa (japonica cultivar-group)]
Length = 254
Score = 151 bits (382), Expect = 5e-36
Identities = 74/87 (85%), Positives = 79/87 (90%)
Frame = -2
Query: 574 EGGFDPNAVATANILESSTPVVDGKQYYVLTVLTRTADGDEGGKHQLIRAAVKDGKLYIC 395
EGGF+ +AVATANILESS PVV GKQYY +TVLTRTADGDEGGKHQLI A V DGKLYIC
Sbjct: 168 EGGFESDAVATANILESSAPVVGGKQYYSVTVLTRTADGDEGGKHQLITATVNDGKLYIC 227
Query: 394 KAQAGDKRWFKGARRYVESTASSFSVA 314
KAQAGDKRWFKGAR++VES ASSFSVA
Sbjct: 228 KAQAGDKRWFKGARKFVESAASSFSVA 254
>sp|O49080|PSBP_FRIAG Oxygen-evolving enhancer protein 2, chloroplast precursor (OEE2)
(23 kDa subunit of oxygen evolving system of photosystem
II) (OEC 23 kDa subunit) (23 kDa thylakoid membrane
protein) gi|2921508|gb|AAC04809.1| photosystem II oxygen
evolving complex protein 2 precursor [Fritillaria
agrestis]
Length = 264
Score = 146 bits (369), Expect = 2e-34
Identities = 69/87 (79%), Positives = 79/87 (90%)
Frame = -2
Query: 574 EGGFDPNAVATANILESSTPVVDGKQYYVLTVLTRTADGDEGGKHQLIRAAVKDGKLYIC 395
EGGFDP++VATANILE STPVV GKQYY ++VLTRTADGDEGGKH +I A V DGKLYIC
Sbjct: 178 EGGFDPDSVATANILEVSTPVVGGKQYYNISVLTRTADGDEGGKHHVISATVTDGKLYIC 237
Query: 394 KAQAGDKRWFKGARRYVESTASSFSVA 314
KAQAGDKRWFKGA+++VEST +SF+VA
Sbjct: 238 KAQAGDKRWFKGAKKFVESTTTSFNVA 264
>sp|Q40407|PSBP_NARPS Oxygen-evolving enhancer protein 2, chloroplast precursor (OEE2)
(23 kDa subunit of oxygen evolving system of photosystem
II) (OEC 23 kDa subunit) (23 kDa thylakoid membrane
protein) gi|2147750|pir||S63532 NAD(P)H-quinone
oxidoreductase, 23K, precursor - Narcissus
pseudonarcissus gi|780273|emb|CAA55393.1| OEC 23kd
protein [Narcissus pseudonarcissus]
Length = 265
Score = 145 bits (367), Expect = 3e-34
Identities = 70/87 (80%), Positives = 76/87 (86%)
Frame = -2
Query: 574 EGGFDPNAVATANILESSTPVVDGKQYYVLTVLTRTADGDEGGKHQLIRAAVKDGKLYIC 395
EGGF+PNAV TANILE+STPVV GK YY + VLTRTADGDEGGKH LI A V DGKLYIC
Sbjct: 179 EGGFEPNAVRTANILETSTPVVGGKDYYSIHVLTRTADGDEGGKHLLITATVSDGKLYIC 238
Query: 394 KAQAGDKRWFKGARRYVESTASSFSVA 314
KAQAGDKRWFKGA+++VES ASSFS A
Sbjct: 239 KAQAGDKRWFKGAKKFVESAASSFSAA 265
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 468,251,459
Number of Sequences: 1393205
Number of extensions: 9352480
Number of successful extensions: 20483
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 19851
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 20444
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 21530810025
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)