Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC017217A_C01 KMC017217A_c01
(569 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
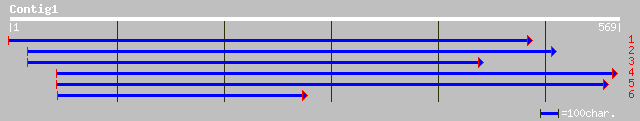
ref|NP_190977.1| fructokinase - like protein; protein id: At3g54... 161 5e-39
dbj|BAB92847.1| putative fructokinase [Oryza sativa (japonica cu... 149 3e-35
gb|AAF27059.1|AC008262_8 F4N2.16 [Arabidopsis thaliana] 69 5e-11
ref|NP_177080.1| fructokinase (Frk1), putative; protein id: At1g... 69 5e-11
gb|AAL26574.1|AF429948_1 putative fructokinase I [Oryza sativa] ... 50 2e-05
>ref|NP_190977.1| fructokinase - like protein; protein id: At3g54090.1, supported by
cDNA: gi_20260523 [Arabidopsis thaliana]
gi|11357296|pir||T47568 fructokinase-like protein -
Arabidopsis thaliana gi|6822055|emb|CAB70983.1|
fructokinase-like protein [Arabidopsis thaliana]
gi|20260524|gb|AAM13160.1| fructokinase-like protein
[Arabidopsis thaliana] gi|22136278|gb|AAM91217.1|
fructokinase-like protein [Arabidopsis thaliana]
Length = 471
Score = 161 bits (408), Expect = 5e-39
Identities = 77/82 (93%), Positives = 81/82 (97%)
Frame = -3
Query: 567 DVLITPYTCDRTGSGDAVVAAILRKLTTCPEMFENQDVLERQLRFAVAAGIIAQWTIGAV 388
DVLITP+TCDRTGSGDAVVA I+RKLTTCPEMFE+QDV+ERQLRFAVAAGIIAQWTIGAV
Sbjct: 390 DVLITPFTCDRTGSGDAVVAGIMRKLTTCPEMFEDQDVMERQLRFAVAAGIIAQWTIGAV 449
Query: 387 RGFPTESATQNLKEQVYVPSMW 322
RGFPTESATQNLKEQVYVPSMW
Sbjct: 450 RGFPTESATQNLKEQVYVPSMW 471
>dbj|BAB92847.1| putative fructokinase [Oryza sativa (japonica cultivar-group)]
Length = 529
Score = 149 bits (375), Expect = 3e-35
Identities = 71/82 (86%), Positives = 76/82 (92%)
Frame = -3
Query: 567 DVLITPYTCDRTGSGDAVVAAILRKLTTCPEMFENQDVLERQLRFAVAAGIIAQWTIGAV 388
D LITPYT DRTGSGDAVVAA +RKLT+CPEM+E+QD LER LRFAVAAGII+QWTIGAV
Sbjct: 448 DALITPYTTDRTGSGDAVVAAAIRKLTSCPEMYEDQDTLERNLRFAVAAGIISQWTIGAV 507
Query: 387 RGFPTESATQNLKEQVYVPSMW 322
RGFPTESA QNLKEQVYVPSMW
Sbjct: 508 RGFPTESAAQNLKEQVYVPSMW 529
>gb|AAF27059.1|AC008262_8 F4N2.16 [Arabidopsis thaliana]
Length = 568
Score = 68.6 bits (166), Expect = 5e-11
Identities = 30/64 (46%), Positives = 42/64 (64%)
Frame = -3
Query: 567 DVLITPYTCDRTGSGDAVVAAILRKLTTCPEMFENQDVLERQLRFAVAAGIIAQWTIGAV 388
DV ITP+T D + SGD +VA ++R LT P++ N+ LER R+A+ GII QW +
Sbjct: 435 DVPITPFTRDMSASGDGIVAGLIRMLTVQPDLMNNKGYLERTARYAIECGIIDQWLLAQT 494
Query: 387 RGFP 376
RG+P
Sbjct: 495 RGYP 498
>ref|NP_177080.1| fructokinase (Frk1), putative; protein id: At1g69200.1 [Arabidopsis
thaliana] gi|25404799|pir||A96716 probable fructokinase
F23O10.21 [imported] - Arabidopsis thaliana
gi|12325093|gb|AAG52502.1|AC018364_20 putative
fructokinase; 80884-78543 [Arabidopsis thaliana]
Length = 614
Score = 68.6 bits (166), Expect = 5e-11
Identities = 30/64 (46%), Positives = 42/64 (64%)
Frame = -3
Query: 567 DVLITPYTCDRTGSGDAVVAAILRKLTTCPEMFENQDVLERQLRFAVAAGIIAQWTIGAV 388
DV ITP+T D + SGD +VA ++R LT P++ N+ LER R+A+ GII QW +
Sbjct: 481 DVPITPFTRDMSASGDGIVAGLIRMLTVQPDLMNNKGYLERTARYAIECGIIDQWLLAQT 540
Query: 387 RGFP 376
RG+P
Sbjct: 541 RGYP 544
>gb|AAL26574.1|AF429948_1 putative fructokinase I [Oryza sativa] gi|20161284|dbj|BAB90210.1|
putative fructokinase I [Oryza sativa (japonica
cultivar-group)] gi|21952837|dbj|BAC06252.1| putative
fructokinase I [Oryza sativa (japonica cultivar-group)]
Length = 323
Score = 50.1 bits (118), Expect = 2e-05
Identities = 26/65 (40%), Positives = 35/65 (53%)
Frame = -3
Query: 540 DRTGSGDAVVAAILRKLTTCPEMFENQDVLERQLRFAVAAGIIAQWTIGAVRGFPTESAT 361
D TG+GDA V A+LR++ P ++Q LE ++FA A G I GA+ PTE
Sbjct: 257 DTTGAGDAFVGALLRRIVQDPSSLQDQKKLEEAIKFANACGAITATKKGAIPSLPTEVEV 316
Query: 360 QNLKE 346
L E
Sbjct: 317 LKLME 321
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 493,136,263
Number of Sequences: 1393205
Number of extensions: 10418861
Number of successful extensions: 23041
Number of sequences better than 10.0: 39
Number of HSP's better than 10.0 without gapping: 22375
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 23034
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20956655091
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)