Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC017077A_C01 KMC017077A_c01
(621 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
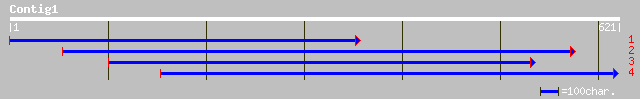
ref|NP_177870.1| unknown protein; protein id: At1g77460.1 [Arabi... 59 4e-08
dbj|BAC42703.1| unknown protein [Arabidopsis thaliana] 59 4e-08
ref|NP_175078.1| hypothetical protein; protein id: At1g44120.1 [... 50 2e-05
ref|XP_213524.1| similar to RIKEN cDNA 1700012B07 [Mus musculus]... 33 3.0
ref|NP_509552.1| Reverse transcriptase and Integrase core domai... 33 3.9
>ref|NP_177870.1| unknown protein; protein id: At1g77460.1 [Arabidopsis thaliana]
gi|25406506|pir||H96803 unknown protein T5M16.5
[imported] - Arabidopsis thaliana
gi|12323397|gb|AAG51678.1|AC010704_22 unknown protein;
15069-22101 [Arabidopsis thaliana]
Length = 2110
Score = 59.3 bits (142), Expect = 4e-08
Identities = 26/42 (61%), Positives = 34/42 (80%)
Frame = -2
Query: 620 IDKVVTDGVYSGLFSLNHDRNNDGSSRTLEIEIVWSNRISND 495
IDKVVT+G YSG SLNH+ + D SSR+L+IEI WSNR +++
Sbjct: 2067 IDKVVTEGEYSGSLSLNHENSKDASSRSLDIEIAWSNRTTDE 2108
>dbj|BAC42703.1| unknown protein [Arabidopsis thaliana]
Length = 434
Score = 59.3 bits (142), Expect = 4e-08
Identities = 26/42 (61%), Positives = 34/42 (80%)
Frame = -2
Query: 620 IDKVVTDGVYSGLFSLNHDRNNDGSSRTLEIEIVWSNRISND 495
IDKVVT+G YSG SLNH+ + D SSR+L+IEI WSNR +++
Sbjct: 391 IDKVVTEGEYSGSLSLNHENSKDASSRSLDIEIAWSNRTTDE 432
>ref|NP_175078.1| hypothetical protein; protein id: At1g44120.1 [Arabidopsis thaliana]
gi|25405158|pir||E96505 hypothetical protein T7O23.25
[imported] - Arabidopsis thaliana
gi|12320824|gb|AAG50555.1|AC074228_10 hypothetical
protein [Arabidopsis thaliana]
Length = 2114
Score = 50.1 bits (118), Expect = 2e-05
Identities = 24/39 (61%), Positives = 31/39 (78%), Gaps = 1/39 (2%)
Frame = -2
Query: 620 IDKVVTDGVYSGLFSLNHDRNNDGSS-RTLEIEIVWSNR 507
IDKV+++G YSG+F LN + D SS R+LEIEIVWSN+
Sbjct: 2074 IDKVLSEGSYSGIFKLNDESKKDNSSDRSLEIEIVWSNQ 2112
>ref|XP_213524.1| similar to RIKEN cDNA 1700012B07 [Mus musculus] [Rattus norvegicus]
Length = 130
Score = 33.1 bits (74), Expect = 3.0
Identities = 14/21 (66%), Positives = 18/21 (85%)
Frame = -3
Query: 292 KEVSPAKENKKKICSELYRLV 230
K++SPAK+N KKIC E+ RLV
Sbjct: 40 KDLSPAKKNYKKICKEMRRLV 60
>ref|NP_509552.1| Reverse transcriptase and Integrase core domain containing protein
family member [Caenorhabditis elegans]
gi|7498548|pir||T15962 hypothetical protein F07C7.1 -
Caenorhabditis elegans gi|1166600|gb|AAA85753.1|
Hypothetical protein F07C7.1 [Caenorhabditis elegans]
Length = 1879
Score = 32.7 bits (73), Expect = 3.9
Identities = 22/85 (25%), Positives = 39/85 (45%)
Frame = +1
Query: 301 LSPSHDINNSIYTYNEETSSASLQNWIFQYVDNL*AKPCPTYEACCRALSMLPSGKAARY 480
+ P +D NN ++ + E ++ YVD + A P Y+ A + LP G+ Y
Sbjct: 1783 IGPDYDTNNPLFHEDGEAEDRPVE-----YVDPITAIPEIAYD---NAETRLPQGRTREY 1834
Query: 481 LSQMSSLEILLDHTISISRVREEPS 555
L + + + + I+RV +PS
Sbjct: 1835 LGRKAKAPYINYNHAEITRVLSDPS 1859
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 539,384,169
Number of Sequences: 1393205
Number of extensions: 11327974
Number of successful extensions: 28833
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 27894
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 28831
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25017613016
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)