Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC016993A_C01 KMC016993A_c01
(589 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
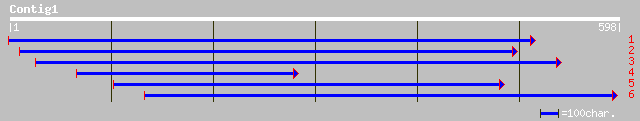
gb|AAM20595.1| integral membrane protein, putative [Arabidopsis ... 163 1e-39
ref|NP_188806.1| integral membrane protein, putative; protein id... 163 1e-39
gb|AAM62936.1| unknown [Arabidopsis thaliana] 158 5e-38
ref|NP_567640.1| putative protein; protein id: At4g21910.1 [Arab... 158 5e-38
ref|NP_564787.1| expressed protein; protein id: At1g61890.1, sup... 155 5e-37
>gb|AAM20595.1| integral membrane protein, putative [Arabidopsis thaliana]
Length = 506
Score = 163 bits (413), Expect = 1e-39
Identities = 75/96 (78%), Positives = 83/96 (86%)
Frame = -2
Query: 588 NGIQPVLSGVAVGCGCQTFVAYVNVGCYYGVGIPLGAVLGFYFKFGAKGIWLGMLGGTVM 409
NGIQPVLSGVAVGCG QTFVA VNVGCYY +GIPLGA+ GFYF FGAKGIW GM+GGTV+
Sbjct: 409 NGIQPVLSGVAVGCGWQTFVAKVNVGCYYIIGIPLGALFGFYFNFGAKGIWTGMIGGTVI 468
Query: 408 QTIILMWVTFRTDWIKEVEEASKRLNKWEEKTEPLL 301
QT IL WVTFRTDW KEVEEASKRL+KW K + ++
Sbjct: 469 QTFILAWVTFRTDWTKEVEEASKRLDKWSNKKQEVV 504
>ref|NP_188806.1| integral membrane protein, putative; protein id: At3g21690.1,
supported by cDNA: gi_20466555 [Arabidopsis thaliana]
gi|11994404|dbj|BAB02363.1|
gb|AAC28507.1~gene_id:MIL23.25~strong similarity to
unknown protein [Arabidopsis thaliana]
Length = 506
Score = 163 bits (413), Expect = 1e-39
Identities = 75/96 (78%), Positives = 83/96 (86%)
Frame = -2
Query: 588 NGIQPVLSGVAVGCGCQTFVAYVNVGCYYGVGIPLGAVLGFYFKFGAKGIWLGMLGGTVM 409
NGIQPVLSGVAVGCG QTFVA VNVGCYY +GIPLGA+ GFYF FGAKGIW GM+GGTV+
Sbjct: 409 NGIQPVLSGVAVGCGWQTFVAKVNVGCYYIIGIPLGALFGFYFNFGAKGIWTGMIGGTVI 468
Query: 408 QTIILMWVTFRTDWIKEVEEASKRLNKWEEKTEPLL 301
QT IL WVTFRTDW KEVEEASKRL+KW K + ++
Sbjct: 469 QTFILAWVTFRTDWTKEVEEASKRLDKWSNKKQEVV 504
>gb|AAM62936.1| unknown [Arabidopsis thaliana]
Length = 507
Score = 158 bits (399), Expect = 5e-38
Identities = 72/97 (74%), Positives = 85/97 (87%)
Frame = -2
Query: 588 NGIQPVLSGVAVGCGCQTFVAYVNVGCYYGVGIPLGAVLGFYFKFGAKGIWLGMLGGTVM 409
NGIQPVLSGVAVGCG QT+VAYVNVGCYY VGIP+G +LGF F F AKGIW GM+GGT+M
Sbjct: 411 NGIQPVLSGVAVGCGWQTYVAYVNVGCYYVVGIPVGCILGFTFDFQAKGIWTGMIGGTLM 470
Query: 408 QTIILMWVTFRTDWIKEVEEASKRLNKWEEKTEPLLN 298
QT+IL++VT+RTDW KEVE+A KRL+ W++K EPL N
Sbjct: 471 QTLILLYVTYRTDWDKEVEKARKRLDLWDDKKEPLQN 507
>ref|NP_567640.1| putative protein; protein id: At4g21910.1 [Arabidopsis thaliana]
gi|15809935|gb|AAL06895.1| AT4g21910/T8O5_120
[Arabidopsis thaliana] gi|22137012|gb|AAM91351.1|
At4g21910/T8O5_120 [Arabidopsis thaliana]
Length = 507
Score = 158 bits (399), Expect = 5e-38
Identities = 72/97 (74%), Positives = 85/97 (87%)
Frame = -2
Query: 588 NGIQPVLSGVAVGCGCQTFVAYVNVGCYYGVGIPLGAVLGFYFKFGAKGIWLGMLGGTVM 409
NGIQPVLSGVAVGCG QT+VAYVNVGCYY VGIP+G +LGF F F AKGIW GM+GGT+M
Sbjct: 411 NGIQPVLSGVAVGCGWQTYVAYVNVGCYYVVGIPVGCILGFTFDFQAKGIWTGMIGGTLM 470
Query: 408 QTIILMWVTFRTDWIKEVEEASKRLNKWEEKTEPLLN 298
QT+IL++VT+RTDW KEVE+A KRL+ W++K EPL N
Sbjct: 471 QTLILLYVTYRTDWDKEVEKARKRLDLWDDKKEPLQN 507
>ref|NP_564787.1| expressed protein; protein id: At1g61890.1, supported by cDNA:
gi_13272458, supported by cDNA: gi_15028186, supported
by cDNA: gi_15081772, supported by cDNA: gi_16209723
[Arabidopsis thaliana] gi|7486693|pir||T02134
hypothetical protein F8K4.9 - Arabidopsis thaliana
gi|3367522|gb|AAC28507.1| EST gb|T04691 comes from this
gene. [Arabidopsis thaliana]
gi|13272459|gb|AAK17168.1|AF325100_1 unknown protein
[Arabidopsis thaliana] gi|16209724|gb|AAL14417.1|
At1g61890/F8K4_9 [Arabidopsis thaliana]
Length = 501
Score = 155 bits (391), Expect = 5e-37
Identities = 73/96 (76%), Positives = 82/96 (85%)
Frame = -2
Query: 588 NGIQPVLSGVAVGCGCQTFVAYVNVGCYYGVGIPLGAVLGFYFKFGAKGIWLGMLGGTVM 409
NGIQPVLSGVAVGCG Q FVAYVN+GCYY VGIP+G VLGF + GAKGIW GM+GGT+M
Sbjct: 404 NGIQPVLSGVAVGCGWQAFVAYVNIGCYYVVGIPVGFVLGFTYDMGAKGIWTGMIGGTLM 463
Query: 408 QTIILMWVTFRTDWIKEVEEASKRLNKWEEKTEPLL 301
QTIIL+ VT RTDW KEVE+AS RL++WEE EPLL
Sbjct: 464 QTIILVIVTLRTDWDKEVEKASSRLDQWEESREPLL 499
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 547,112,448
Number of Sequences: 1393205
Number of extensions: 12261438
Number of successful extensions: 33053
Number of sequences better than 10.0: 179
Number of HSP's better than 10.0 without gapping: 31530
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 32979
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 22283372436
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)