Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC016941A_C01 KMC016941A_c01
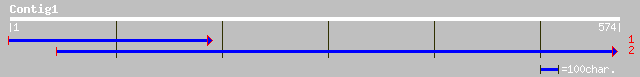
(574 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||T52404 hypothetical protein MMB12.23 [imported] - Arabidops... 88 7e-17
ref|NP_175433.1| hypothetical protein; protein id: At1g50140.1 [... 82 6e-15
pir||G96537 hypothetical protein F2J10.1 [imported] - Arabidopsi... 82 6e-15
ref|NP_194529.1| putative protein; protein id: At4g28000.1 [Arab... 36 0.30
ref|NP_188608.1| ATPase, putative; protein id: At3g19740.1 [Arab... 36 0.39
>pir||T52404 hypothetical protein MMB12.23 [imported] - Arabidopsis thaliana
gi|9294441|dbj|BAB02561.1| gene_id:MMB12.23~unknown
protein [Arabidopsis thaliana]
Length = 208
Score = 88.2 bits (217), Expect = 7e-17
Identities = 54/126 (42%), Positives = 77/126 (60%), Gaps = 27/126 (21%)
Frame = -3
Query: 572 YFPDG---LHKSGPKSIQSEFFQKVQEKFELLSGPIVLICGQNQAQ--SKGQLLFSM--- 417
YFPD L ++ PKS Q+EF KVQE F+ LS P+V+ICG+N+ + SK + F+M
Sbjct: 48 YFPDSSQWLSRAVPKSKQNEFVDKVQEMFDKLSSPVVMICGRNKIETGSKEREKFTMILP 107
Query: 416 ------------------NKGQKTSKDGEIYKPF-NVLSIQPPKDEKLLVIFKKXLEEDR 294
G+KTS+D EIYK F NV+++ PPK+E+ L++F K L EDR
Sbjct: 108 NFGRLAKLPLPLKRLTEGLTGRKTSEDNEIYKLFTNVMNLVPPKEEENLIVFNKQLGEDR 167
Query: 293 KIVISQ 276
+IV+S+
Sbjct: 168 RIVMSR 173
>ref|NP_175433.1| hypothetical protein; protein id: At1g50140.1 [Arabidopsis
thaliana]
Length = 640
Score = 81.6 bits (200), Expect = 6e-15
Identities = 53/126 (42%), Positives = 73/126 (57%), Gaps = 27/126 (21%)
Frame = -3
Query: 572 YFPDG---LHKSGPKSIQSEFFQKVQEKFELLSGPIVLICGQNQAQ--SKGQLLFSM--- 417
YFPD L ++ PK+ + EF KV+E F+ LSGPIV+ICGQN+ + SK + F+M
Sbjct: 111 YFPDSTQWLSRAVPKTRRKEFVDKVKEMFDKLSGPIVMICGQNKIETGSKEREKFTMVLP 170
Query: 416 ------------------NKGQKTSKDGEIYKPF-NVLSIQPPKDEKLLVIFKKXLEEDR 294
G+ S++ EIYK F NV+ + PPK+E L +FKK L EDR
Sbjct: 171 NLSRVVKLPLPLKGLTEGFTGRGKSEENEIYKLFTNVMRLHPPKEEDTLRLFKKQLGEDR 230
Query: 293 KIVISQ 276
+IVIS+
Sbjct: 231 RIVISR 236
>pir||G96537 hypothetical protein F2J10.1 [imported] - Arabidopsis thaliana
gi|8569089|gb|AAF76434.1|AC015445_1 Contains similarity
to p60 katanin from Chlamydomonas reinhardtii
gb|AF205377 and contains an AAA domain PF|00004.
[Arabidopsis thaliana]
Length = 627
Score = 81.6 bits (200), Expect = 6e-15
Identities = 53/126 (42%), Positives = 73/126 (57%), Gaps = 27/126 (21%)
Frame = -3
Query: 572 YFPDG---LHKSGPKSIQSEFFQKVQEKFELLSGPIVLICGQNQAQ--SKGQLLFSM--- 417
YFPD L ++ PK+ + EF KV+E F+ LSGPIV+ICGQN+ + SK + F+M
Sbjct: 111 YFPDSTQWLSRAVPKTRRKEFVDKVKEMFDKLSGPIVMICGQNKIETGSKEREKFTMVLP 170
Query: 416 ------------------NKGQKTSKDGEIYKPF-NVLSIQPPKDEKLLVIFKKXLEEDR 294
G+ S++ EIYK F NV+ + PPK+E L +FKK L EDR
Sbjct: 171 NLSRVVKLPLPLKGLTEGFTGRGKSEENEIYKLFTNVMRLHPPKEEDTLRLFKKQLGEDR 230
Query: 293 KIVISQ 276
+IVIS+
Sbjct: 231 RIVISR 236
>ref|NP_194529.1| putative protein; protein id: At4g28000.1 [Arabidopsis thaliana]
gi|7487959|pir||T02901 MSP1 protein homolog T13J8.110 -
Arabidopsis thaliana gi|4455359|emb|CAB36769.1| putative
protein [Arabidopsis thaliana]
gi|7269654|emb|CAB79602.1| putative protein [Arabidopsis
thaliana]
Length = 726
Score = 36.2 bits (82), Expect = 0.30
Identities = 26/86 (30%), Positives = 44/86 (50%), Gaps = 1/86 (1%)
Frame = -3
Query: 539 KSIQSEFFQKV-QEKFELLSGPIVLICGQNQAQSKGQLLFSMNKGQKTSKDGEIYKPFNV 363
K QSE F K+ Q LSGP++++ +LL + Q+ + P+N+
Sbjct: 214 KLCQSERFYKLFQRLLTKLSGPVLVL--------GSRLLEPEDDCQEVGEGISALFPYNI 265
Query: 362 LSIQPPKDEKLLVIFKKXLEEDRKIV 285
I+PP+DE L+ +K E+D K++
Sbjct: 266 -EIRPPEDENQLMSWKTRFEDDMKVI 290
>ref|NP_188608.1| ATPase, putative; protein id: At3g19740.1 [Arabidopsis thaliana]
Length = 439
Score = 35.8 bits (81), Expect = 0.39
Identities = 14/29 (48%), Positives = 24/29 (82%)
Frame = -3
Query: 362 LSIQPPKDEKLLVIFKKXLEEDRKIVISQ 276
+++ PPK+E+ L++F K L EDR+IV+S+
Sbjct: 1 MNLVPPKEEENLIVFNKQLGEDRRIVMSR 29
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 410,664,107
Number of Sequences: 1393205
Number of extensions: 7496763
Number of successful extensions: 20017
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 19777
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 20012
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 21243732558
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)