Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC016901A_C01 KMC016901A_c01
(536 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
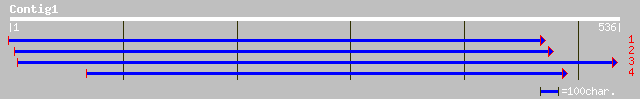
gb|AAM97321.1| homeodomain protein GhHOX1 [Gossypium hirsutum] 117 9e-26
gb|AAK19610.1|AF336277_1 BNLGHi8377 [Gossypium hirsutum] 117 9e-26
sp|P46607|HGL2_ARATH Homeobox protein GLABRA2 (Homeobox-leucine ... 89 3e-17
ref|NP_565223.1| homeodomain protein, GLABRA2 (GL2); protein id:... 89 3e-17
gb|AAL83725.1|AF328842_1 homeodomain protein HB2 [Picea abies] 74 1e-12
>gb|AAM97321.1| homeodomain protein GhHOX1 [Gossypium hirsutum]
Length = 753
Score = 117 bits (293), Expect = 9e-26
Identities = 58/71 (81%), Positives = 67/71 (93%), Gaps = 1/71 (1%)
Frame = -1
Query: 536 PDGLESRPLVITTRKEEKN-TEGGSLFTIAFQILTNASPTAKLTVESVDSVNTLVSCTLR 360
PDGLESRPLVI++R E+ N TEGGSL T+AFQILTN+SPTAKLT+ESV+SVNT+VSCTLR
Sbjct: 683 PDGLESRPLVISSRHEKSNDTEGGSLLTVAFQILTNSSPTAKLTMESVESVNTIVSCTLR 742
Query: 359 NIRTSLQCEDG 327
NI+TSLQCEDG
Sbjct: 743 NIKTSLQCEDG 753
>gb|AAK19610.1|AF336277_1 BNLGHi8377 [Gossypium hirsutum]
Length = 758
Score = 117 bits (293), Expect = 9e-26
Identities = 58/71 (81%), Positives = 67/71 (93%), Gaps = 1/71 (1%)
Frame = -1
Query: 536 PDGLESRPLVITTRKEEKN-TEGGSLFTIAFQILTNASPTAKLTVESVDSVNTLVSCTLR 360
PDGLESRPLVI++R E+ N TEGGSL T+AFQILTN+SPTAKLT+ESV+SVNT+VSCTLR
Sbjct: 688 PDGLESRPLVISSRHEKSNDTEGGSLLTVAFQILTNSSPTAKLTMESVESVNTIVSCTLR 747
Query: 359 NIRTSLQCEDG 327
NI+TSLQCEDG
Sbjct: 748 NIKTSLQCEDG 758
>sp|P46607|HGL2_ARATH Homeobox protein GLABRA2 (Homeobox-leucine zipper protein ATHB-10)
(HD-ZIP protein ATHB-10) gi|25386229|pir||D96829
homeobox protein (GLABRA2), 66648-63167 [imported] -
Arabidopsis thaliana gi|1695244|gb|AAC80260.1|
homeodomain protein [Arabidopsis thaliana]
gi|12324584|gb|AAG52245.1|AC011717_13 homeobox protein
(GLABRA2); 66648-63167 [Arabidopsis thaliana]
Length = 745
Score = 89.0 bits (219), Expect = 3e-17
Identities = 43/69 (62%), Positives = 55/69 (79%)
Frame = -1
Query: 536 PDGLESRPLVITTRKEEKNTEGGSLFTIAFQILTNASPTAKLTVESVDSVNTLVSCTLRN 357
PDG+ESRPLVIT+ ++++N++GGSL T+A Q L N SP AKL +ESV+SV LVS TL N
Sbjct: 676 PDGVESRPLVITSTQDDRNSQGGSLLTLALQTLINPSPAAKLNMESVESVTNLVSVTLHN 735
Query: 356 IRTSLQCED 330
I+ SLQ ED
Sbjct: 736 IKRSLQIED 744
>ref|NP_565223.1| homeodomain protein, GLABRA2 (GL2); protein id: At1g79840.1,
supported by cDNA: gi_13430763 [Arabidopsis thaliana]
gi|2129609|pir||S71478 homeotic protein Athb-10 -
Arabidopsis thaliana gi|1212757|emb|CAA91183.1| HD-ZIP
[Arabidopsis thaliana]
gi|13430764|gb|AAK26004.1|AF360294_1 putative homeobox
protein GLABRA2 [Arabidopsis thaliana]
gi|20152552|emb|CAD29714.1| homeodomain-leucine zipper
10 [Arabidopsis thaliana] gi|25054963|gb|AAN71955.1|
putative homeobox protein GLABRA2 [Arabidopsis thaliana]
Length = 747
Score = 89.0 bits (219), Expect = 3e-17
Identities = 43/69 (62%), Positives = 55/69 (79%)
Frame = -1
Query: 536 PDGLESRPLVITTRKEEKNTEGGSLFTIAFQILTNASPTAKLTVESVDSVNTLVSCTLRN 357
PDG+ESRPLVIT+ ++++N++GGSL T+A Q L N SP AKL +ESV+SV LVS TL N
Sbjct: 678 PDGVESRPLVITSTQDDRNSQGGSLLTLALQTLINPSPAAKLNMESVESVTNLVSVTLHN 737
Query: 356 IRTSLQCED 330
I+ SLQ ED
Sbjct: 738 IKRSLQIED 746
>gb|AAL83725.1|AF328842_1 homeodomain protein HB2 [Picea abies]
Length = 708
Score = 73.9 bits (180), Expect = 1e-12
Identities = 37/73 (50%), Positives = 49/73 (66%), Gaps = 4/73 (5%)
Frame = -1
Query: 536 PDGLESRPLVITTRKEEKNTE----GGSLFTIAFQILTNASPTAKLTVESVDSVNTLVSC 369
P+G + RPL + GGSL T+AFQIL N+ PTAKLTVESV++VN L+SC
Sbjct: 635 PNGPKCRPLALNPSGNGVGVNSPRVGGSLLTVAFQILVNSLPTAKLTVESVETVNNLISC 694
Query: 368 TLRNIRTSLQCED 330
T++ I+ +L CED
Sbjct: 695 TVQKIKAALHCED 707
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 441,203,229
Number of Sequences: 1393205
Number of extensions: 8916731
Number of successful extensions: 19519
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 19043
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 19498
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 18173652336
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)