Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
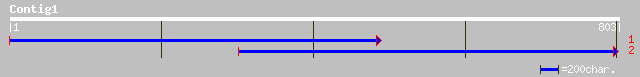
Query= KMC016810A_C01 KMC016810A_c01
(803 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_507483.1| Potassium channel protein family member (51.2 k... 36 0.75
ref|NP_507803.1| Putative cytoplasmic protein family member, nem... 35 1.7
ref|NP_213069.1| putative protein [Aquifex aeolicus] gi|14916731... 33 6.4
ref|NP_114042.1|ND2_15649 NADH dehydrogenase subunit 2 [Mertensi... 32 8.3
>ref|NP_507483.1| Potassium channel protein family member (51.2 kD) [Caenorhabditis
elegans] gi|7509066|pir||T26229 hypothetical protein
W06D12.2 - Caenorhabditis elegans
gi|3880547|emb|CAB04923.1| Hypothetical protein W06D12.2
[Caenorhabditis elegans]
Length = 444
Score = 35.8 bits (81), Expect = 0.75
Identities = 23/70 (32%), Positives = 37/70 (52%)
Frame = +3
Query: 144 FHYQMSVALFTNRRKPLAILSFQTMKP*PPKTDLTWINLSSKIPSNSHMAKKMEIRSYMS 323
F +SVA +R+K + + F ++ P KT T N++ K N M K+ +R +MS
Sbjct: 318 FKNTLSVAAEESRKKSIGVSEFGSIDPSKKKTMRTDGNVAEKYSENMEMGNKLLMR-FMS 376
Query: 324 SSQYKKENEQ 353
+ Q K NE+
Sbjct: 377 NHQKKMLNEK 386
>ref|NP_507803.1| Putative cytoplasmic protein family member, nematode specific
[Caenorhabditis elegans] gi|7509826|pir||T26877
hypothetical protein Y43F8C.3 - Caenorhabditis elegans
gi|3979965|emb|CAA21618.1| Hypothetical protein Y43F8C.3
[Caenorhabditis elegans]
Length = 603
Score = 34.7 bits (78), Expect = 1.7
Identities = 18/52 (34%), Positives = 25/52 (47%)
Frame = -2
Query: 322 DMYDLISIFFAMWELEGILDDRFIHVRSVLGGHGFIVWNDKIANGFLLLVKR 167
D+ D+ IF WELE DR IH+ + GF +W + F L+ R
Sbjct: 185 DLVDIFGIFQNRWELEA---DRMIHLIPIARKFGFDIWGKSLNREFSQLMHR 233
>ref|NP_213069.1| putative protein [Aquifex aeolicus]
gi|14916731|sp|O66510|Y106_AQUAE Hypothetical protein
AQ_106 gi|7517365|pir||C70310 hypothetical protein
aq_106 - Aquifex aeolicus gi|2982842|gb|AAC06464.1|
putative protein [Aquifex aeolicus VF5]
Length = 239
Score = 32.7 bits (73), Expect = 6.4
Identities = 24/74 (32%), Positives = 33/74 (44%)
Frame = +2
Query: 470 TPIL*FTCSSLLILLHHGLCKPANKFENSFYPLIYLQFLAYSVDQQSTFQIKYLYPAHFH 649
+P L F L +L H + E Y L+Y FL Y QQS+ + LY
Sbjct: 133 SPSLSFFLLILYLLQHIFVLFKLFAVEYVRYELVYFLFLEYRTRQQSSRVLVVLYLPEHE 192
Query: 650 KTKKKVILISRQVT 691
K +K V+ +S VT
Sbjct: 193 KFRKSVLNLSVSVT 206
>ref|NP_114042.1|ND2_15649 NADH dehydrogenase subunit 2 [Mertensiella luschani]
gi|13873054|gb|AAK43347.1|AF154053_2 NADH dehydrogenase
subunit 2 [Mertensiella luschani]
Length = 345
Score = 32.3 bits (72), Expect = 8.3
Identities = 32/134 (23%), Positives = 57/134 (41%), Gaps = 13/134 (9%)
Frame = -3
Query: 540 FAGLQRPWWRRISKELQVN*RMGVLKLICWLAKTMDIVGFFI--------FKHVIYVSIT 385
+ GL + R+I + +G + +I + T+ ++ F I F +I + T
Sbjct: 166 WGGLNQVQMRKIMAYSSIA-HLGWMIMILTFSPTLTVLNFLIYIILTTSMFMMMISLLST 224
Query: 384 RTSNG*FSWEAAHSLFYIVNWTCMIL-----SPSFLPCGN*KVSWMIDLSMSDQFWEVMV 220
+ SW +L ++ T M L + FLP W+I M++Q +M
Sbjct: 225 NINKLAVSWSKTPTLSALMMITLMALGGLPPTSGFLP------KWLILQEMTNQHMSLMS 278
Query: 219 SLFGMIRLLMVFFY 178
++ M LL +FFY
Sbjct: 279 TIMAMSTLLSLFFY 292
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 696,727,981
Number of Sequences: 1393205
Number of extensions: 15454149
Number of successful extensions: 30080
Number of sequences better than 10.0: 8
Number of HSP's better than 10.0 without gapping: 28967
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 30064
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 40896270532
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)