Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC016496A_C01 KMC016496A_c01
(621 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
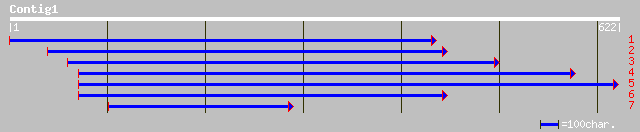
gb|AAO42228.1| unknown protein [Arabidopsis thaliana] 96 3e-19
ref|NP_201148.1| putative protein; protein id: At5g63430.1 [Arab... 96 3e-19
ref|NP_172802.1| DNA binding protein GT-1, putative; protein id:... 43 0.003
gb|AAA66473.1| GT-1 43 0.003
gb|AAG09542.1|AC011810_1 DNA binding protein GT-1 [Arabidopsis t... 43 0.003
>gb|AAO42228.1| unknown protein [Arabidopsis thaliana]
Length = 911
Score = 96.3 bits (238), Expect = 3e-19
Identities = 48/73 (65%), Positives = 56/73 (75%), Gaps = 1/73 (1%)
Frame = -3
Query: 613 RFQVVKGRMALWEEISQNLLADGISRSPGQCKSLWTSLVLKYEEIKNTDGNK-SWQYRED 437
RFQVVKGRMALWEEIS NL A+GI+RSPGQCKSLW SL+ KYEE K + +K SW + ED
Sbjct: 839 RFQVVKGRMALWEEISSNLSAEGINRSPGQCKSLWASLIQKYEESKADERSKTSWPHFED 898
Query: 436 MERAMSNEETPAT 398
M +S TPA+
Sbjct: 899 MNNILSELGTPAS 911
>ref|NP_201148.1| putative protein; protein id: At5g63430.1 [Arabidopsis thaliana]
gi|9758284|dbj|BAB08808.1|
gene_id:MLE2.6~pir||T01257~similar to unknown protein
[Arabidopsis thaliana]
Length = 330
Score = 96.3 bits (238), Expect = 3e-19
Identities = 48/73 (65%), Positives = 56/73 (75%), Gaps = 1/73 (1%)
Frame = -3
Query: 613 RFQVVKGRMALWEEISQNLLADGISRSPGQCKSLWTSLVLKYEEIKNTDGNK-SWQYRED 437
RFQVVKGRMALWEEIS NL A+GI+RSPGQCKSLW SL+ KYEE K + +K SW + ED
Sbjct: 258 RFQVVKGRMALWEEISSNLSAEGINRSPGQCKSLWASLIQKYEESKADERSKTSWPHFED 317
Query: 436 MERAMSNEETPAT 398
M +S TPA+
Sbjct: 318 MNNILSELGTPAS 330
>ref|NP_172802.1| DNA binding protein GT-1, putative; protein id: At1g13450.1
[Arabidopsis thaliana]
Length = 413
Score = 43.1 bits (100), Expect = 0.003
Identities = 25/75 (33%), Positives = 38/75 (50%), Gaps = 6/75 (8%)
Frame = -3
Query: 610 FQVVKGRMALWEEISQNLLADGISRSPGQCKSLWTSLVLKYEEIKNTD-GNKS-----WQ 449
F K LWE+IS + G RSP C W +L+ ++++ K+ D GN S ++
Sbjct: 106 FNTSKSNKHLWEQISSKMREKGFDRSPTMCTDKWRNLLKEFKKAKHHDRGNGSAKMSYYK 165
Query: 448 YREDMERAMSNEETP 404
ED+ R S + TP
Sbjct: 166 EIEDILRERSKKVTP 180
>gb|AAA66473.1| GT-1
Length = 406
Score = 43.1 bits (100), Expect = 0.003
Identities = 25/75 (33%), Positives = 38/75 (50%), Gaps = 6/75 (8%)
Frame = -3
Query: 610 FQVVKGRMALWEEISQNLLADGISRSPGQCKSLWTSLVLKYEEIKNTD-GNKS-----WQ 449
F K LWE+IS + G RSP C W +L+ ++++ K+ D GN S ++
Sbjct: 106 FNTSKSNKHLWEQISSKMREKGFDRSPTMCTDKWRNLLKEFKKAKHHDRGNGSAKMSYYK 165
Query: 448 YREDMERAMSNEETP 404
ED+ R S + TP
Sbjct: 166 EIEDILRERSKKVTP 180
>gb|AAG09542.1|AC011810_1 DNA binding protein GT-1 [Arabidopsis thaliana]
Length = 406
Score = 43.1 bits (100), Expect = 0.003
Identities = 25/75 (33%), Positives = 38/75 (50%), Gaps = 6/75 (8%)
Frame = -3
Query: 610 FQVVKGRMALWEEISQNLLADGISRSPGQCKSLWTSLVLKYEEIKNTD-GNKS-----WQ 449
F K LWE+IS + G RSP C W +L+ ++++ K+ D GN S ++
Sbjct: 106 FNTSKSNKHLWEQISSKMREKGFDRSPTMCTDKWRNLLKEFKKAKHHDRGNGSAKMSYYK 165
Query: 448 YREDMERAMSNEETP 404
ED+ R S + TP
Sbjct: 166 EIEDILRERSKKVTP 180
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 518,905,159
Number of Sequences: 1393205
Number of extensions: 11200478
Number of successful extensions: 26693
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 25962
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26681
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25017613016
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)