Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC016489A_C01 KMC016489A_c01
(625 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
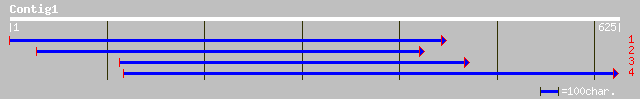
ref|NP_197530.1| putative protein; protein id: At5g20300.1 [Arab... 65 7e-10
gb|AAM20511.1| putative chloroplast outer membrane protein [Arab... 51 1e-05
ref|NP_179255.1| putative chloroplast outer membrane protein; pr... 51 1e-05
gb|AAA53276.1| GTP-binding protein 50 3e-05
pir||S49910 chloroplast outer envelope protein OEP86 precursor -... 50 3e-05
>ref|NP_197530.1| putative protein; protein id: At5g20300.1 [Arabidopsis thaliana]
Length = 787
Score = 65.1 bits (157), Expect = 7e-10
Identities = 36/87 (41%), Positives = 58/87 (66%)
Frame = -3
Query: 620 KEMVLSGSLQSEFRLSRSLRASVGADLNSRKMGQISVKMSSSEHLQIALVGVFSIFKVLS 441
+E+VL+ LQ++FR +R V ++N+RKMG+I+VK++SSEH +IAL+ ++FK L
Sbjct: 706 RELVLNYGLQTQFRPARGTNIDVNINMNNRKMGKINVKLNSSEHWEIALISALTMFKALV 765
Query: 440 RRKPTSKNVMKEVMDRD*VMFVDFVLS 360
RR SK M E + + V+F++S
Sbjct: 766 RR---SKTEMTEENEEE--KIVNFLVS 787
>gb|AAM20511.1| putative chloroplast outer membrane protein [Arabidopsis thaliana]
Length = 1202
Score = 51.2 bits (121), Expect = 1e-05
Identities = 23/58 (39%), Positives = 40/58 (68%)
Frame = -3
Query: 617 EMVLSGSLQSEFRLSRSLRASVGADLNSRKMGQISVKMSSSEHLQIALVGVFSIFKVL 444
++ + G++QS+ + RS A+LN+R GQ+SV+++SSE LQ+A+V + +FK L
Sbjct: 1137 DLAIGGNIQSQVPIGRSSNLIARANLNNRGAGQVSVRVNSSEQLQLAMVAIVPLFKKL 1194
>ref|NP_179255.1| putative chloroplast outer membrane protein; protein id: At2g16640.1,
supported by cDNA: gi_20466387 [Arabidopsis thaliana]
gi|25411728|pir||D84542 probable chloroplast outer
membrane protein [imported] - Arabidopsis thaliana
gi|4581108|gb|AAD24598.1| putative chloroplast outer
membrane protein [Arabidopsis thaliana]
Length = 1206
Score = 51.2 bits (121), Expect = 1e-05
Identities = 23/58 (39%), Positives = 40/58 (68%)
Frame = -3
Query: 617 EMVLSGSLQSEFRLSRSLRASVGADLNSRKMGQISVKMSSSEHLQIALVGVFSIFKVL 444
++ + G++QS+ + RS A+LN+R GQ+SV+++SSE LQ+A+V + +FK L
Sbjct: 1137 DLAIGGNIQSQVPIGRSSNLIARANLNNRGAGQVSVRVNSSEQLQLAMVAIVPLFKKL 1194
>gb|AAA53276.1| GTP-binding protein
Length = 879
Score = 49.7 bits (117), Expect = 3e-05
Identities = 23/58 (39%), Positives = 38/58 (64%)
Frame = -3
Query: 617 EMVLSGSLQSEFRLSRSLRASVGADLNSRKMGQISVKMSSSEHLQIALVGVFSIFKVL 444
++ L + QS+ L RS + +V A LN++ GQI+V+ SSS+ LQIAL+ + + K +
Sbjct: 807 DLALGANFQSQISLGRSYKMAVRAGLNNKLSGQINVRTSSSDQLQIALIAILPVAKAI 864
>pir||S49910 chloroplast outer envelope protein OEP86 precursor - garden pea
gi|599958|emb|CAA83453.1| chloroplast outer envelope
protein 86 [Pisum sativum]
Length = 879
Score = 49.7 bits (117), Expect = 3e-05
Identities = 23/58 (39%), Positives = 38/58 (64%)
Frame = -3
Query: 617 EMVLSGSLQSEFRLSRSLRASVGADLNSRKMGQISVKMSSSEHLQIALVGVFSIFKVL 444
++ L + QS+ L RS + +V A LN++ GQI+V+ SSS+ LQIAL+ + + K +
Sbjct: 807 DLALGANFQSQISLGRSYKMAVRAGLNNKLSGQINVRTSSSDQLQIALIAILPVAKAI 864
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 511,839,883
Number of Sequences: 1393205
Number of extensions: 10539572
Number of successful extensions: 17258
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 16804
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 17251
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25301904073
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)