Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC016419A_C01 KMC016419A_c01
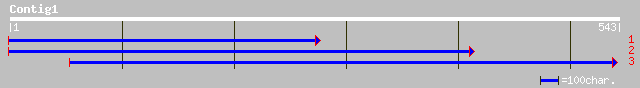
(543 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAB39155.1| SCARECROW [Pisum sativum] 43 0.002
ref|NP_477602.1| wsv080 [shrimp white spot syndrome virus] gi|17... 33 2.2
gb|AAG13663.1|AF263457_1 SCARECROW [Zea mays] 32 3.8
ref|NP_577733.1| hypothetical protein [Pyrococcus furiosus DSM 3... 32 5.0
gb|ZP_00039543.1| hypothetical protein [Xylella fastidiosa Dixon] 32 6.5
>dbj|BAB39155.1| SCARECROW [Pisum sativum]
Length = 819
Score = 43.1 bits (100), Expect = 0.002
Identities = 19/25 (76%), Positives = 21/25 (84%), Gaps = 3/25 (12%)
Frame = -2
Query: 542 KDLCLLTASAWRPPF---TAIPHHH 477
KDLCLLTASAWRPP+ T IPHH+
Sbjct: 795 KDLCLLTASAWRPPYHTNTIIPHHN 819
>ref|NP_477602.1| wsv080 [shrimp white spot syndrome virus]
gi|17016478|gb|AAL33084.1| wsv080 [shrimp white spot
syndrome virus] gi|19481729|gb|AAL89005.1| WSSV137
[shrimp white spot syndrome virus]
Length = 129
Score = 33.1 bits (74), Expect = 2.2
Identities = 26/86 (30%), Positives = 41/86 (47%), Gaps = 12/86 (13%)
Frame = -1
Query: 441 VMALSYESILGFTFRSSRISSAPISAL*VTSPR----INSPSNVKIHCL--------SKY 298
++AL +E+ L FTF PI++L V R I SPS + CL ++Y
Sbjct: 9 LLALHFEAYLRFTFNFLEHKLLPITSLTVVWNRLPLDIQSPSLFSVSCLLSLRAESSARY 68
Query: 297 ALILILFSIPLFLSLE*EIVTCLQSM 220
L + L+S + SL ++T S+
Sbjct: 69 LLAISLYSSSILSSLHIYLLTSTHSL 94
>gb|AAG13663.1|AF263457_1 SCARECROW [Zea mays]
Length = 668
Score = 32.3 bits (72), Expect = 3.8
Identities = 13/13 (100%), Positives = 13/13 (100%)
Frame = -2
Query: 542 KDLCLLTASAWRP 504
KDLCLLTASAWRP
Sbjct: 649 KDLCLLTASAWRP 661
>ref|NP_577733.1| hypothetical protein [Pyrococcus furiosus DSM 3638]
gi|18891900|gb|AAL80128.1| hypothetical protein
[Pyrococcus furiosus DSM 3638]
Length = 257
Score = 32.0 bits (71), Expect = 5.0
Identities = 19/61 (31%), Positives = 31/61 (50%), Gaps = 3/61 (4%)
Frame = +1
Query: 13 IIKLLIHQGYTNTHKSQNIIVTSVLSSLNKILSLNMYHFS---HFDITFFYLTYILFHVI 183
+I L GY + S ++ + LS LN ILSL +Y +S F + F +T + + +
Sbjct: 176 LISLTFLLGYLQVNTSLDVSLLVFLSPLNSILSLLIYGYSGILSFSVEFLLVTSLFYITV 235
Query: 184 L 186
L
Sbjct: 236 L 236
>gb|ZP_00039543.1| hypothetical protein [Xylella fastidiosa Dixon]
Length = 586
Score = 31.6 bits (70), Expect = 6.5
Identities = 24/73 (32%), Positives = 38/73 (51%)
Frame = -1
Query: 534 LLAHCFSLETSLHCHSPSS*LVSSYAKLSDVVMALSYESILGFTFRSSRISSAPISAL*V 355
L AH L+ S H + S+ LVS A S+++ ++ S + RS+ + + L +
Sbjct: 105 LYAHLIGLDASFHDRNRSNELVSRLATDSELLRSI-IGSTMSVATRSTVTAIGSLIMLFI 163
Query: 354 TSPRINSPSNVKI 316
TSPR+ S S V I
Sbjct: 164 TSPRLASLSLVVI 176
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 442,352,753
Number of Sequences: 1393205
Number of extensions: 8959973
Number of successful extensions: 18985
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 18453
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 18976
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 18750593680
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)