Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
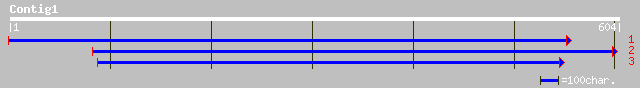
Query= KMC016368A_C01 KMC016368A_c01
(604 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM97011.1| expressed protein [Arabidopsis thaliana] gi|23197... 202 4e-51
ref|NP_563815.1| expressed protein; protein id: At1g08380.1, sup... 202 4e-51
emb|CAB75430.1| putative 16kDa membrane protein [Nicotiana tabacum] 192 3e-48
ref|NP_691016.1| hypothetical protein [Oceanobacillus iheyensis ... 34 1.3
ref|NP_508131.1| Predicted CDS, homeobox protein [Caenorhabditis... 33 3.7
>gb|AAM97011.1| expressed protein [Arabidopsis thaliana] gi|23197966|gb|AAN15510.1|
expressed protein [Arabidopsis thaliana]
Length = 140
Score = 202 bits (513), Expect = 4e-51
Identities = 94/118 (79%), Positives = 106/118 (89%)
Frame = -1
Query: 577 TSDFVKSPVSARNPLRQAVAMGNGGRVKCFERNWLRTDFNVIGFGLIGWIAPSSIPAIDG 398
+S F+K +SA+NPLR A A +GGRV CFERNWLR D NV+GFGLIGW+APSSIPAI+G
Sbjct: 25 SSSFLKPTLSAKNPLRLAGA--SGGRVTCFERNWLRRDLNVVGFGLIGWLAPSSIPAING 82
Query: 397 KSLTGLFFDSIGTELAHFPSPPALDSQFWLWLVTWHLGLFIVLTFGQIGFKGRTDEYF 224
KSLTGLFFDSIGTELAHFP+PPAL SQFWLWLVTWHLGLF+ LTFGQIGFKGRT++YF
Sbjct: 83 KSLTGLFFDSIGTELAHFPTPPALTSQFWLWLVTWHLGLFLCLTFGQIGFKGRTEDYF 140
>ref|NP_563815.1| expressed protein; protein id: At1g08380.1, supported by cDNA:
34623., supported by cDNA: gi_15293052, supported by
cDNA: gi_20259136 [Arabidopsis thaliana]
gi|15293053|gb|AAK93637.1| unknown protein [Arabidopsis
thaliana] gi|20259137|gb|AAM14284.1| unknown protein
[Arabidopsis thaliana] gi|21592969|gb|AAM64918.1|
putative 16kDa membrane protein [Arabidopsis thaliana]
gi|21702253|emb|CAD37939.1| photosystem I subunit O
[Arabidopsis thaliana]
Length = 140
Score = 202 bits (513), Expect = 4e-51
Identities = 94/118 (79%), Positives = 106/118 (89%)
Frame = -1
Query: 577 TSDFVKSPVSARNPLRQAVAMGNGGRVKCFERNWLRTDFNVIGFGLIGWIAPSSIPAIDG 398
+S F+K +SA+NPLR A A +GGRV CFERNWLR D NV+GFGLIGW+APSSIPAI+G
Sbjct: 25 SSSFLKPTLSAKNPLRLAGA--SGGRVTCFERNWLRRDLNVVGFGLIGWLAPSSIPAING 82
Query: 397 KSLTGLFFDSIGTELAHFPSPPALDSQFWLWLVTWHLGLFIVLTFGQIGFKGRTDEYF 224
KSLTGLFFDSIGTELAHFP+PPAL SQFWLWLVTWHLGLF+ LTFGQIGFKGRT++YF
Sbjct: 83 KSLTGLFFDSIGTELAHFPTPPALTSQFWLWLVTWHLGLFLCLTFGQIGFKGRTEDYF 140
>emb|CAB75430.1| putative 16kDa membrane protein [Nicotiana tabacum]
Length = 143
Score = 192 bits (488), Expect = 3e-48
Identities = 92/126 (73%), Positives = 107/126 (84%), Gaps = 1/126 (0%)
Frame = -1
Query: 598 SPASSRLTSDFVKSPVSARNPLRQAVAMGNGGRVKC-FERNWLRTDFNVIGFGLIGWIAP 422
SP + L+S +KSPV+ARNPLR VA +GG+ C F+R+WLR D NVIGFGLIGW+AP
Sbjct: 18 SPKKACLSSGLLKSPVTARNPLR--VAQASGGKFTCNFQRDWLRRDLNVIGFGLIGWLAP 75
Query: 421 SSIPAIDGKSLTGLFFDSIGTELAHFPSPPALDSQFWLWLVTWHLGLFIVLTFGQIGFKG 242
SSIP I+GKSL+GLFFDSIGTEL+HFP+ PAL SQFWLWLV WHLGLFI LTFGQIGFKG
Sbjct: 76 SSIPVINGKSLSGLFFDSIGTELSHFPTGPALTSQFWLWLVCWHLGLFICLTFGQIGFKG 135
Query: 241 RTDEYF 224
RT++YF
Sbjct: 136 RTEDYF 141
>ref|NP_691016.1| hypothetical protein [Oceanobacillus iheyensis HTE831]
gi|22775773|dbj|BAC12051.1| hypothetical conserved
protein [Oceanobacillus iheyensis]
Length = 364
Score = 34.3 bits (77), Expect = 1.3
Identities = 29/93 (31%), Positives = 49/93 (52%), Gaps = 6/93 (6%)
Frame = -1
Query: 505 GRVKCFERNWLRTDFNVIGFGLIGWIAPS--SIPAIDGKSLTGLFFDSIGT----ELAHF 344
G + F ++L D+ V G + W+ + IPA+D LFF SIG +A+F
Sbjct: 48 GAIIFFILSYLLVDYIV---GFLRWVEDALIKIPAMD------LFFGSIGLIVGLVIAYF 98
Query: 343 PSPPALDSQFWLWLVTWHLGLFIVLTFGQIGFK 245
+ P D+ + +++ L FI++TFG +GF+
Sbjct: 99 INIPLQDTG--IHILSQVLPFFIMITFGYLGFQ 129
>ref|NP_508131.1| Predicted CDS, homeobox protein [Caenorhabditis elegans]
gi|7506106|pir||T29183 hypothetical protein M6.3 -
Caenorhabditis elegans gi|1293798|gb|AAA98706.1|
Hypothetical protein M6.3 [Caenorhabditis elegans]
Length = 257
Score = 32.7 bits (73), Expect = 3.7
Identities = 16/44 (36%), Positives = 24/44 (54%)
Frame = +3
Query: 435 PINPNPITLKSVRSQFLSKHFTLPPFPIATACLKGFLALTGDFT 566
P P P+ + S F++ FTLPPF +A++ + G GD T
Sbjct: 12 PYLPAPLCMSSRAIAFIAPFFTLPPFQLASSLVWGLKIDFGDTT 55
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 529,856,355
Number of Sequences: 1393205
Number of extensions: 12043672
Number of successful extensions: 37470
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 35864
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 37420
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23711793746
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)