Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC016335A_C01 KMC016335A_c01
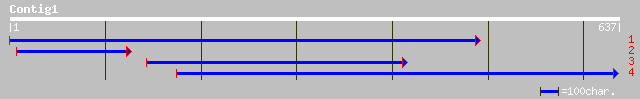
(636 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAG34812.1|AF243377_1 glutathione S-transferase GST 22 [Glyci... 167 1e-40
gb|AAM34480.1| putative glutathione S-transferase [Phaseolus acu... 131 7e-30
gb|AAG34814.1|AF243379_1 glutathione S-transferase GST 24 [Glyci... 131 7e-30
ref|NP_180643.1| glutathione transferase, putative; protein id: ... 126 2e-28
emb|CAA72973.1| glutathione transferase [Arabidopsis thaliana] 122 5e-27
>gb|AAG34812.1|AF243377_1 glutathione S-transferase GST 22 [Glycine max]
Length = 210
Score = 167 bits (423), Expect = 1e-40
Identities = 77/94 (81%), Positives = 87/94 (91%)
Frame = -1
Query: 630 LEGLPSDPKLIEESDEKVPKVLDIYEDRLSKSQYLAGDFFSLADLSPLPFGHYLVNPTGR 451
L G PSD K+IEESD+K+ KVLD+YE+RLSKS+YLAGDFFSLADLS LPFGHYLVN TGR
Sbjct: 117 LTGAPSDQKVIEESDKKIEKVLDVYEERLSKSKYLAGDFFSLADLSHLPFGHYLVNQTGR 176
Query: 450 GDFVRERKHVSAWWDDISSRPSWQKVLQLYKYPV 349
G+ VR+RKHVSAWWDDIS+RP+WQKVLQLYKYPV
Sbjct: 177 GNLVRDRKHVSAWWDDISNRPAWQKVLQLYKYPV 210
>gb|AAM34480.1| putative glutathione S-transferase [Phaseolus acutifolius]
Length = 215
Score = 131 bits (330), Expect = 7e-30
Identities = 62/95 (65%), Positives = 75/95 (78%)
Frame = -1
Query: 633 ALEGLPSDPKLIEESDEKVPKVLDIYEDRLSKSQYLAGDFFSLADLSPLPFGHYLVNPTG 454
+L G+ DPK+IEES+ K+ KVLD+YEDRLSK +YL GDF SLAD+S LPF Y+VN
Sbjct: 121 SLFGITPDPKVIEESEAKLLKVLDVYEDRLSKGKYLGGDFLSLADISHLPFIDYIVNKMN 180
Query: 453 RGDFVRERKHVSAWWDDISSRPSWQKVLQLYKYPV 349
+G ++ERKHVS WWDDISSRPSW+KV QLY PV
Sbjct: 181 KGYLIKERKHVSDWWDDISSRPSWKKVNQLYPPPV 215
>gb|AAG34814.1|AF243379_1 glutathione S-transferase GST 24 [Glycine max]
Length = 215
Score = 131 bits (330), Expect = 7e-30
Identities = 61/95 (64%), Positives = 78/95 (81%)
Frame = -1
Query: 633 ALEGLPSDPKLIEESDEKVPKVLDIYEDRLSKSQYLAGDFFSLADLSPLPFGHYLVNPTG 454
+L G+ DPK+IEES+ K+ +VL+IYE+RLSK++YLAGDFFS+AD+S LPF Y+VN G
Sbjct: 121 SLFGVTPDPKVIEESEAKLVQVLNIYEERLSKTKYLAGDFFSIADISHLPFLDYVVNNMG 180
Query: 453 RGDFVRERKHVSAWWDDISSRPSWQKVLQLYKYPV 349
+ + ERKHV AWWDDISSRPSW KVLQLY+ P+
Sbjct: 181 KKYLLEERKHVGAWWDDISSRPSWNKVLQLYRAPI 215
>ref|NP_180643.1| glutathione transferase, putative; protein id: At2g30860.1,
supported by cDNA: gi_13926309 [Arabidopsis thaliana]
gi|25287119|pir||E84713 glutathione S-transferase
[imported] - Arabidopsis thaliana
gi|3201613|gb|AAC20720.1| glutathione S-transferase
[Arabidopsis thaliana]
gi|13926310|gb|AAK49621.1|AF372905_1 At2g30860/F7F1.7
[Arabidopsis thaliana] gi|27363352|gb|AAO11595.1|
At2g30860/F7F1.7 [Arabidopsis thaliana]
Length = 215
Score = 126 bits (317), Expect = 2e-28
Identities = 57/91 (62%), Positives = 72/91 (78%)
Frame = -1
Query: 624 GLPSDPKLIEESDEKVPKVLDIYEDRLSKSQYLAGDFFSLADLSPLPFGHYLVNPTGRGD 445
G PSD KLI+ES+EK+ VLD+YE LSKS+YLAGDF SLADL+ LPF YLV P G+
Sbjct: 124 GFPSDEKLIKESEEKLAGVLDVYEAHLSKSKYLAGDFVSLADLAHLPFTDYLVGPIGKAY 183
Query: 444 FVRERKHVSAWWDDISSRPSWQKVLQLYKYP 352
+++RKHVSAWWDDISSRP+W++ + Y +P
Sbjct: 184 MIKDRKHVSAWWDDISSRPAWKETVAKYSFP 214
>emb|CAA72973.1| glutathione transferase [Arabidopsis thaliana]
Length = 215
Score = 122 bits (305), Expect = 5e-27
Identities = 54/91 (59%), Positives = 72/91 (78%)
Frame = -1
Query: 624 GLPSDPKLIEESDEKVPKVLDIYEDRLSKSQYLAGDFFSLADLSPLPFGHYLVNPTGRGD 445
G PSD KLI+ES+EK+ VLD+Y+ + +KS+YLAGDF SLADL+ LPF YLV P G+
Sbjct: 124 GFPSDEKLIKESEEKLAGVLDVYKAQRAKSKYLAGDFVSLADLAHLPFTDYLVGPIGKAY 183
Query: 444 FVRERKHVSAWWDDISSRPSWQKVLQLYKYP 352
+++RKHVSAWWDDISSRP+W++ + Y +P
Sbjct: 184 MIKDRKHVSAWWDDISSRPAWKETVAKYSFP 214
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 526,549,706
Number of Sequences: 1393205
Number of extensions: 11238570
Number of successful extensions: 30455
Number of sequences better than 10.0: 229
Number of HSP's better than 10.0 without gapping: 29241
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 30340
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 26439068301
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)