Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC016310A_C01 KMC016310A_c01
(1297 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
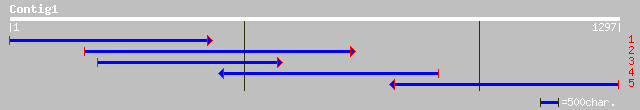
ref|NP_186829.1| putative ribosome recycling factor; protein id:... 292 9e-78
emb|CAD41132.1| OSJNBa0084K20.12 [Oryza sativa (japonica cultiva... 284 2e-75
ref|NP_243290.1| ribosome recycling factor [Bacillus halodurans]... 136 6e-31
gb|ZP_00082204.1| hypothetical protein [Geobacter metallireducens] 132 9e-30
gb|ZP_00094055.1| hypothetical protein [Novosphingobium aromatic... 130 4e-29
>ref|NP_186829.1| putative ribosome recycling factor; protein id: At3g01800.1
[Arabidopsis thaliana]
gi|6091742|gb|AAF03454.1|AC010797_30 putative ribosome
recycling factor [Arabidopsis thaliana]
Length = 284
Score = 292 bits (747), Expect = 9e-78
Identities = 159/286 (55%), Positives = 206/286 (71%), Gaps = 26/286 (9%)
Frame = -3
Query: 1151 MSGYIRRAISCQNLLQ-LRNSA-------FHVCDVPGAIQIRALESNRVSTLPAAANFIV 996
M+ ++RRA+S +N + LR S+ F P + + + ++P +F+
Sbjct: 1 MAMFVRRALSSRNAIGFLRRSSVVVQSRDFSFISSPNLMMMGGRDLQFDLSMP---DFLR 57
Query: 995 ESRRAFAKGRKSKDDAGMSTIEVPLVLGPTIKANAVSQMEAAISALSTELSKLRTGRASP 816
+SRR FAKG+KSKDD+G ++ +GPT+KA A SQMEAAI ALS +L+KLRTGRA+P
Sbjct: 58 DSRRGFAKGKKSKDDSGTGMVDAAPDIGPTVKAAASSQMEAAIDALSRDLTKLRTGRAAP 117
Query: 815 GMLDHIIVETGGVKIPLVRIALVSVIDPKTLSVNPYDPQTLKQLENAIVSSPLGLNPKAD 636
GMLDHI+VETGGVK+PL +ALVSV+DPKTLSVNPYDP T+K+LE AIV+SPLGLNPK D
Sbjct: 118 GMLDHIVVETGGVKMPLNHLALVSVLDPKTLSVNPYDPDTVKELEKAIVASPLGLNPKLD 177
Query: 635 GERLIAAIP------------------PLTKEHMQAMNKLVTKSCEDARQSIRRARQKAM 510
G+RL+A+IP P+ +QAM K+VTKS E +QSIRRARQKA+
Sbjct: 178 GQRLVASIPAYESSSLSNFISLRLSLDPIYLYELQAMCKIVTKSSEVVKQSIRRARQKAL 237
Query: 509 DAIKKLNSSLPKDDIKRLEKEVDDLTKKFIKTAEDTCKAKEKEISQ 372
D IKK SSLPKD++KRLEKEVD+LTKKF+K+AED CK+KEKEI++
Sbjct: 238 DTIKKAGSSLPKDEVKRLEKEVDELTKKFVKSAEDMCKSKEKEITE 283
>emb|CAD41132.1| OSJNBa0084K20.12 [Oryza sativa (japonica cultivar-group)]
Length = 260
Score = 284 bits (727), Expect = 2e-75
Identities = 142/210 (67%), Positives = 176/210 (83%)
Frame = -3
Query: 1004 FIVESRRAFAKGRKSKDDAGMSTIEVPLVLGPTIKANAVSQMEAAISALSTELSKLRTGR 825
F++E+RR FAKG+KSKDD T++ +GPT+K+ A QMEAA+ ALS ELSKLRTGR
Sbjct: 49 FLLEARRGFAKGKKSKDDGRGDTVQDAPDIGPTVKSAATQQMEAAVVALSRELSKLRTGR 108
Query: 824 ASPGMLDHIIVETGGVKIPLVRIALVSVIDPKTLSVNPYDPQTLKQLENAIVSSPLGLNP 645
ASPGMLDHI+VET GVK+ L R+A+VSV+D TLSV PYDP ++K +E+AI+SSPLG+NP
Sbjct: 109 ASPGMLDHIMVETTGVKVALNRLAVVSVLDAHTLSVMPYDPSSMKSIEHAIISSPLGINP 168
Query: 644 KADGERLIAAIPPLTKEHMQAMNKLVTKSCEDARQSIRRARQKAMDAIKKLNSSLPKDDI 465
DG R+IA IPPLTKE++QA+ K+VTKS ED +QSIRRARQKA+D IKK S +PKDD+
Sbjct: 169 TPDGNRIIANIPPLTKENIQALCKVVTKSAEDFKQSIRRARQKALDTIKKSASGMPKDDV 228
Query: 464 KRLEKEVDDLTKKFIKTAEDTCKAKEKEIS 375
KRLEKEV++LTKKFIK+A+D CKAKEKEIS
Sbjct: 229 KRLEKEVEELTKKFIKSADDMCKAKEKEIS 258
>ref|NP_243290.1| ribosome recycling factor [Bacillus halodurans]
gi|14195173|sp|Q9KA66|RRF_BACHD Ribosome recycling
factor gi|25299708|pir||H83952 ribosome recycling factor
frr [imported] - Bacillus halodurans (strain C-125)
gi|10175044|dbj|BAB06143.1| ribosome recycling factor
[Bacillus halodurans]
Length = 185
Score = 136 bits (343), Expect = 6e-31
Identities = 73/177 (41%), Positives = 110/177 (61%), Gaps = 2/177 (1%)
Frame = -3
Query: 896 NAVSQMEAAISALSTELSKLRTGRASPGMLDHIIVETGGVKIPLVRIALVSVIDPKTLSV 717
+A +M A AL EL+KLR GRA+P MLD I VE G + PL ++A +SV + + L +
Sbjct: 8 DAEQRMTKATEALGRELAKLRAGRANPAMLDRITVEYYGAETPLNQLATISVPEARLLVI 67
Query: 716 NPYDPQTLKQLENAIVSSPLGLNPKADGERLIAAIPPLTKEHMQAMNKLVTKSCEDARQS 537
P+D ++ +E AI S LGL P DG + IPPLT+E + + KLV KS E+A+ +
Sbjct: 68 QPFDKSSISDIERAIQKSDLGLTPSNDGTVIRITIPPLTEERRRDLTKLVKKSAEEAKVA 127
Query: 536 IRRARQKAMDAIKK--LNSSLPKDDIKRLEKEVDDLTKKFIKTAEDTCKAKEKEISQ 372
+R R+ A D +KK + L +DD++R+ ++V LT K+I+ + +AKEKEI +
Sbjct: 128 VRNIRRDANDDLKKRQKDGELTEDDLRRVTEDVQKLTDKYIEQIDQKAEAKEKEIME 184
>gb|ZP_00082204.1| hypothetical protein [Geobacter metallireducens]
Length = 185
Score = 132 bits (333), Expect = 9e-30
Identities = 66/176 (37%), Positives = 112/176 (63%), Gaps = 2/176 (1%)
Frame = -3
Query: 899 ANAVSQMEAAISALSTELSKLRTGRASPGMLDHIIVETGGVKIPLVRIALVSVIDPKTLS 720
A+ + ME ++ AL E K+RTGRA+ G+LD I VE G PL ++A +SV +P+T++
Sbjct: 7 ADMKAHMEKSVDALRREYQKVRTGRATTGLLDDIKVEYYGNPSPLSQVATLSVPEPRTIT 66
Query: 719 VNPYDPQTLKQLENAIVSSPLGLNPKADGERLIAAIPPLTKEHMQAMNKLVTKSCEDARQ 540
+ P++ + + +E AI+++ LG P DG+ + ++PPLT+E + + K + K ED +
Sbjct: 67 IQPWEAKLIPVIEKAIMNANLGFTPANDGKTIRISLPPLTEERRKEIVKTLKKMAEDTKV 126
Query: 539 SIRRARQKAMDAIKKL--NSSLPKDDIKRLEKEVDDLTKKFIKTAEDTCKAKEKEI 378
++R R+ +D++KKL + + +DD+KR EKEV D+T F+ E+ KEKE+
Sbjct: 127 AVRNIRRDGIDSLKKLEKDKKISEDDLKRAEKEVQDVTNTFVAKVEEVLTHKEKEV 182
>gb|ZP_00094055.1| hypothetical protein [Novosphingobium aromaticivorans]
Length = 185
Score = 130 bits (328), Expect = 4e-29
Identities = 74/177 (41%), Positives = 109/177 (60%), Gaps = 2/177 (1%)
Frame = -3
Query: 902 KANAVSQMEAAISALSTELSKLRTGRASPGMLDHIIVETGGVKIPLVRIALVSVIDPKTL 723
KA+ +M+ A+ +L +LS LRTGRA+ +LD I+VE G +PL ++A VS +P+ L
Sbjct: 6 KADLERRMKGAVESLRGDLSGLRTGRANTALLDPIVVEVYGSHMPLAQVATVSAPEPRLL 65
Query: 722 SVNPYDPQTLKQLENAIVSSPLGLNPKADGERLIAAIPPLTKEHMQAMNKLVTKSCEDAR 543
SV +D + +E AI S+ LGLNP +DG L IP LT+E + + KL +K E+AR
Sbjct: 66 SVQVWDKSNVTPVEKAIRSAGLGLNPISDGNTLRLPIPDLTEERRKELAKLASKYAENAR 125
Query: 542 QSIRRARQKAMDAIK--KLNSSLPKDDIKRLEKEVDDLTKKFIKTAEDTCKAKEKEI 378
+IR R+ MDA+K + + +D+ KR E ++ LT IK A++ AKEKEI
Sbjct: 126 IAIRNVRRDGMDALKADESKKEISEDERKRAETDLQKLTDDIIKQADEAAAAKEKEI 182
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,099,727,197
Number of Sequences: 1393205
Number of extensions: 24225391
Number of successful extensions: 89864
Number of sequences better than 10.0: 195
Number of HSP's better than 10.0 without gapping: 78377
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 88914
length of database: 448,689,247
effective HSP length: 126
effective length of database: 273,145,417
effective search space used: 83309352185
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)