Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC016258A_C01 KMC016258A_c01
(1107 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
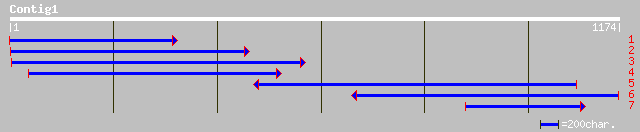
ref|NP_201487.1| putative protein; protein id: At5g66860.1, supp... 294 1e-78
ref|NP_194093.1| putative protein; protein id: At4g23620.1, supp... 168 1e-40
dbj|BAB92303.1| P0451D05.2 [Oryza sativa (japonica cultivar-group)] 130 4e-29
gb|ZP_00009136.1| hypothetical protein [Rhodopseudomonas palustris] 82 1e-14
ref|NP_229427.1| general stress protein Ctc [Thermotoga maritima... 77 5e-13
>ref|NP_201487.1| putative protein; protein id: At5g66860.1, supported by cDNA:
gi_15450929, supported by cDNA: gi_17978762 [Arabidopsis
thaliana] gi|9758136|dbj|BAB08628.1|
gene_id:MUD21.12~pir||T05594~similar to unknown protein
[Arabidopsis thaliana] gi|15450930|gb|AAK96736.1| Unknown
protein [Arabidopsis thaliana] gi|17978763|gb|AAL47375.1|
unknown protein [Arabidopsis thaliana]
Length = 249
Score = 294 bits (753), Expect = 1e-78
Identities = 150/228 (65%), Positives = 178/228 (77%), Gaps = 4/228 (1%)
Frame = -2
Query: 1085 TAAASRLSSPAFQRG----HSYHTIQAIPREHSGSGVAARDRMQGRIPAVVFLQDLLSKD 918
TA + L + +F R H Y TIQAIPRE +G GV+ARDR GRIPAVVF Q LL D
Sbjct: 12 TAEVADLPASSFGRSIRCIHQYQTIQAIPREATGRGVSARDRTIGRIPAVVFPQSLLDTD 71
Query: 917 AGNRSAAKKHLLTVEKKQIKAILNSVQTSFFCSTRFPLQIRAGSGSSHLVESGTVLPVKI 738
A R ++K LLT +KKQIK+I++SV FFCST F LQIRAG GSS LVESG VLP+K+
Sbjct: 72 ASKRGVSRKQLLTADKKQIKSIIDSVGLPFFCSTTFQLQIRAGQGSSTLVESGRVLPLKV 131
Query: 737 HMDQESGQILNLVFVWAEEGMNLKVDVPVVFKGEDVCPGLQKGGFLNKIRTSLKFLCPSE 558
H D+E+G+ILNLVFVWA++G LKVDVPVVFKG D CPGLQKGG L IR++LK L P+E
Sbjct: 132 HRDEETGKILNLVFVWADDGEKLKVDVPVVFKGLDHCPGLQKGGNLRTIRSTLKLLGPAE 191
Query: 557 HIPSKIEVDVSNLDIEDRIFMRDIEVHPSLKLLSKNENMPICKVVPTS 414
HIPSKIEVDVSNLDIED++ ++D+ HPSLKLLSKNE MP+CK+V TS
Sbjct: 192 HIPSKIEVDVSNLDIEDKVLLQDVVFHPSLKLLSKNETMPVCKIVATS 239
>ref|NP_194093.1| putative protein; protein id: At4g23620.1, supported by cDNA: 6527.,
supported by cDNA: gi_18253004 [Arabidopsis thaliana]
gi|7486750|pir||T05594 hypothetical protein F9D16.90 -
Arabidopsis thaliana gi|4454031|emb|CAA23028.1| putative
protein [Arabidopsis thaliana] gi|7269210|emb|CAB79317.1|
putative protein [Arabidopsis thaliana]
gi|18253005|gb|AAL62429.1| putative protein [Arabidopsis
thaliana] gi|21389687|gb|AAM48042.1| putative protein
[Arabidopsis thaliana] gi|21594031|gb|AAM65949.1| unknown
[Arabidopsis thaliana]
Length = 264
Score = 168 bits (426), Expect = 1e-40
Identities = 90/213 (42%), Positives = 135/213 (63%), Gaps = 2/213 (0%)
Frame = -2
Query: 1058 PAFQRGHSYH--TIQAIPREHSGSGVAARDRMQGRIPAVVFLQDLLSKDAGNRSAAKKHL 885
P F R H TI A+PR SG ++A++R GR+P+++F Q+ + K L
Sbjct: 41 PGFPRPDPKHAETILAVPRSVSGKSISAKERKAGRVPSIIFEQE------DGQHGGNKRL 94
Query: 884 LTVEKKQIKAILNSVQTSFFCSTRFPLQIRAGSGSSHLVESGTVLPVKIHMDQESGQILN 705
++V+ QI+ ++N + SFF S F +++RA GS ++E LP IH+ + LN
Sbjct: 95 ISVQTNQIRKLVNHLGYSFFLSRLFDVEVRAEIGSDEVIEKVRALPRAIHLHSGTDAPLN 154
Query: 704 LVFVWAEEGMNLKVDVPVVFKGEDVCPGLQKGGFLNKIRTSLKFLCPSEHIPSKIEVDVS 525
+ F+ A G LKVD+P+VF G+DV PGL+KG LN I+ ++KFLCP+E IP IEVD+S
Sbjct: 155 VTFIRAPPGALLKVDIPLVFIGDDVSPGLKKGASLNTIKRTVKFLCPAEIIPPYIEVDLS 214
Query: 524 NLDIEDRIFMRDIEVHPSLKLLSKNENMPICKV 426
LDI ++ M D++VHP+LKL+ K+++ PI KV
Sbjct: 215 QLDIGQKLVMGDLKVHPALKLI-KSKDEPIVKV 246
>dbj|BAB92303.1| P0451D05.2 [Oryza sativa (japonica cultivar-group)]
Length = 393
Score = 130 bits (327), Expect = 4e-29
Identities = 70/167 (41%), Positives = 109/167 (64%), Gaps = 1/167 (0%)
Frame = -2
Query: 1025 IQAIPREHSGSGVAARDRMQGRIPAVVFLQDLLSKDAGNRSAAKKHLLTVEKKQIKAILN 846
I A+PR SG VAA++R GR+PA+VF Q+ ++ GN K L++V+ KQI+ +++
Sbjct: 66 ILAVPRASSGRHVAAKERKAGRVPAIVFEQEN-GQEGGN-----KRLVSVQSKQIRKLVD 119
Query: 845 SVQTSFFCSTRFPLQIRAG-SGSSHLVESGTVLPVKIHMDQESGQILNLVFVWAEEGMNL 669
+ SFF S F LQ+ + +G L+ES VLP K+H+ + + LN+ F+ A L
Sbjct: 120 HLGRSFFLSRLFRLQVWSEHAGQGELIESVRVLPRKVHLHAGTDEPLNVTFMRAPSSALL 179
Query: 668 KVDVPVVFKGEDVCPGLQKGGFLNKIRTSLKFLCPSEHIPSKIEVDV 528
K+DVP++F GED PGL+KG + N I+ ++K+LCP++ +P IEVD+
Sbjct: 180 KIDVPLMFIGEDASPGLRKGAYFNTIKRTVKYLCPADIVPPYIEVDL 226
>gb|ZP_00009136.1| hypothetical protein [Rhodopseudomonas palustris]
Length = 230
Score = 82.0 bits (201), Expect = 1e-14
Identities = 58/208 (27%), Positives = 106/208 (50%)
Frame = -2
Query: 1025 IQAIPREHSGSGVAARDRMQGRIPAVVFLQDLLSKDAGNRSAAKKHLLTVEKKQIKAILN 846
++A R SG G A +R GR+P V++ N+S ++VE+K+++
Sbjct: 7 LKATARPKSGKGAARAERRAGRVPGVIY--------GDNQSPLP---ISVEEKELRL--- 52
Query: 845 SVQTSFFCSTRFPLQIRAGSGSSHLVESGTVLPVKIHMDQESGQILNLVFVWAEEGMNLK 666
+ F +T F + + G H V+P H+D +++ F+ G ++
Sbjct: 53 RILAGRFLTTVFDVVL---DGKKH-----RVIPRDYHLDPVRDFPIHVDFLRLGAGATIR 104
Query: 665 VDVPVVFKGEDVCPGLQKGGFLNKIRTSLKFLCPSEHIPSKIEVDVSNLDIEDRIFMRDI 486
V VP+ KG +V PG+++GG N + +++ P+E+IP IE DVS LDI + + DI
Sbjct: 105 VSVPLHLKGLEVAPGVKRGGTFNIVTHTVELEAPAENIPQFIEADVSTLDIGVSLHLSDI 164
Query: 485 EVHPSLKLLSKNENMPICKVVPTSLGNQ 402
+ +K +S+ +++ + +VP S N+
Sbjct: 165 ALPTGVKSVSR-DDVTLVTIVPPSGYNE 191
>ref|NP_229427.1| general stress protein Ctc [Thermotoga maritima]
gi|7674238|sp|Q9X1W2|RL25_THEMA Probable 50S ribosomal
protein L25 gi|7462395|pir||C72229 general stress protein
Ctc - Thermotoga maritima (strain MSB8)
gi|4982200|gb|AAD36694.1|AE001806_4 general stress
protein Ctc [Thermotoga maritima]
Length = 215
Score = 77.0 bits (188), Expect = 5e-13
Identities = 54/214 (25%), Positives = 103/214 (47%)
Frame = -2
Query: 1028 TIQAIPREHSGSGVAARDRMQGRIPAVVFLQDLLSKDAGNRSAAKKHLLTVEKKQIKAIL 849
+++A RE G A R R +G +PAVV+ A + + +++ ++ I
Sbjct: 3 SLEARVREVKGKREARRLRRRGEVPAVVY-----------GPATEPIPVKIKRSVLEKIF 51
Query: 848 NSVQTSFFCSTRFPLQIRAGSGSSHLVESGTVLPVKIHMDQESGQILNLVFVWAEEGMNL 669
+++ S P+Q+ + V TV + D+ S +++L F +G +
Sbjct: 52 HTI------SEATPIQLIIKDDQGNTVAEKTVFLKMVQRDKVSETVVHLDFYEPTKGHRM 105
Query: 668 KVDVPVVFKGEDVCPGLQKGGFLNKIRTSLKFLCPSEHIPSKIEVDVSNLDIEDRIFMRD 489
+++VP+ G+ V G++KGGFL + + +P +IEVDVS+LD+ D I RD
Sbjct: 106 RINVPLKVVGKPV--GVEKGGFLEVFHEEIPVETDPDKVPQEIEVDVSSLDLGDVIHARD 163
Query: 488 IEVHPSLKLLSKNENMPICKVVPTSLGNQAPVDE 387
+++ +K L + E + +VP + + +E
Sbjct: 164 LKLPEGVKCLLEEEEAVVSVLVPKEVAIEEATEE 197
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 978,094,142
Number of Sequences: 1393205
Number of extensions: 22308114
Number of successful extensions: 58759
Number of sequences better than 10.0: 104
Number of HSP's better than 10.0 without gapping: 55031
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 58602
length of database: 448,689,247
effective HSP length: 125
effective length of database: 274,538,622
effective search space used: 66712885146
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)