Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC016208A_C01 KMC016208A_c01
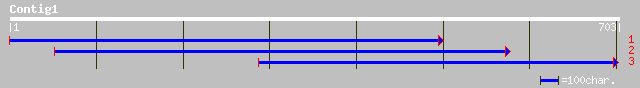
(703 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAA32122.1| cytochrome c oxidase III gi|447158|prf||1913438A ... 38 0.16
gb|AAA63544.1| cytochrome c oxidase III 34 2.3
sp|P14546|COX3_LEITA Cytochrome c oxidase polypeptide III gi|839... 33 3.0
gb|AAA31877.1| cytochrome oxidase 33 3.0
pir||A28782 cytochrome-c oxidase (EC 1.9.3.1) chain III - Trypan... 33 3.9
>gb|AAA32122.1| cytochrome c oxidase III gi|447158|prf||1913438A cytochrome c
oxidase
Length = 288
Score = 37.7 bits (86), Expect = 0.16
Identities = 35/122 (28%), Positives = 49/122 (39%), Gaps = 18/122 (14%)
Frame = -3
Query: 584 CVLF*CLWISIWLFAVTISFRLVLFEL*L---------------FCCL---YGRNWS*FF 459
C+ + LW F + ++F +VLF L L FCCL YG + +F
Sbjct: 85 CIQYCFLWFLCSEFVLFMAFFVVLFGLCLFLCCEFAFVFCLPYMFCCLLCDYGFVFYWYF 144
Query: 458 LSFS*CFFI*CHIVVSVSSFS*GAFLMFVILCRWFVTSRVGLR**FCCCASCLFFTLYII 279
L ++ VS G F+ F +LC WF FCCC C L ++
Sbjct: 145 LDLFNLLINTFYLFVS------GLFVNFFVLCFWFRF--------FCCC--CFVLWLSLL 188
Query: 278 FG 273
FG
Sbjct: 189 FG 190
>gb|AAA63544.1| cytochrome c oxidase III
Length = 111
Score = 33.9 bits (76), Expect = 2.3
Identities = 32/113 (28%), Positives = 44/113 (38%), Gaps = 18/113 (15%)
Frame = -3
Query: 584 CVLF*CLWISIWLFAVTISFRLVLFEL*L---------------FCCL---YGRNWS*FF 459
C+ + LW F + ++F +VLF L L FCCL YG + +F
Sbjct: 13 CIQYCFLWFLCSEFVLFMAFFVVLFGLCLFLCCEFAFVFCLPYMFCCLLCDYGFVFYWYF 72
Query: 458 LSFS*CFFI*CHIVVSVSSFS*GAFLMFVILCRWFVTSRVGLR**FCCCASCL 300
L ++ VS G F+ F +LC WF FCCC L
Sbjct: 73 LDLFNLLINTFYLFVS------GLFVNFFVLCFWFRF--------FCCCCFVL 111
>sp|P14546|COX3_LEITA Cytochrome c oxidase polypeptide III gi|83980|pir||G22848
cytochrome-c oxidase (EC 1.9.3.1) chain III - Leishmania
tarentolae mitochondrion
Length = 284
Score = 33.5 bits (75), Expect = 3.0
Identities = 18/42 (42%), Positives = 27/42 (63%)
Frame = -2
Query: 396 LRRLFNVCDFVQMVRHQQGWFEIIVLLLCFMPVFYAVHNFRL 271
LR LF+VC F++ +++ WF II LL F+ +FY V + L
Sbjct: 69 LRGLFDVCCFIRCIQYCFVWF-IISELLLFLSLFYVVFSLVL 109
>gb|AAA31877.1| cytochrome oxidase
Length = 287
Score = 33.5 bits (75), Expect = 3.0
Identities = 18/42 (42%), Positives = 27/42 (63%)
Frame = -2
Query: 396 LRRLFNVCDFVQMVRHQQGWFEIIVLLLCFMPVFYAVHNFRL 271
LR LF+VC F++ +++ WF II LL F+ +FY V + L
Sbjct: 72 LRGLFDVCCFIRCIQYCFVWF-IISELLLFLSLFYVVFSLVL 112
>pir||A28782 cytochrome-c oxidase (EC 1.9.3.1) chain III - Trypanosoma brucei
mitochondrion (fragment)
Length = 181
Score = 33.1 bits (74), Expect = 3.9
Identities = 25/79 (31%), Positives = 34/79 (42%), Gaps = 3/79 (3%)
Frame = -3
Query: 500 LFCCL---YGRNWS*FFLSFS*CFFI*CHIVVSVSSFS*GAFLMFVILCRWFVTSRVGLR 330
+FCCL YG + +FL ++ VS G F+ F +LC WF
Sbjct: 21 MFCCLLCDYGFVFYWYFLDLFNLLINTFYLFVS------GLFVNFFVLCFWFRF------ 68
Query: 329 **FCCCASCLFFTLYIIFG 273
FCCC C L ++FG
Sbjct: 69 --FCCC--CFVLWLSLLFG 83
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 535,378,216
Number of Sequences: 1393205
Number of extensions: 10450054
Number of successful extensions: 26119
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 25637
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26104
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 32250355128
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)