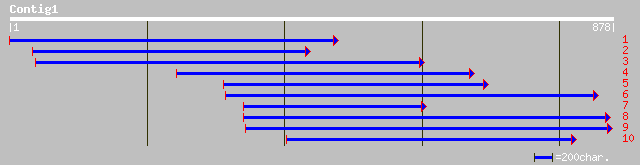
Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC016195A_C01 KMC016195A_c01
(876 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_187867.1| DNA-damage-repair/toleration protein, putative ... 181 2e-44
emb|CAD56505.1| polygalacturonase inhibitor-like protein [Cicer ... 179 4e-44
ref|NP_188718.1| disease resistance protein family (LRR); protei... 177 2e-43
gb|AAM65656.1| leucine rich repeat protein, putative [Arabidopsi... 165 7e-40
gb|AAK64162.1| unknown protein [Arabidopsis thaliana] 165 7e-40
>ref|NP_187867.1| DNA-damage-repair/toleration protein, putative (DRT100); protein
id: At3g12610.1 [Arabidopsis thaliana]
gi|20178285|sp|Q00874|D100_ARATH
DNA-damage-repair/toleration protein DRT100 precursor
gi|9294355|dbj|BAB02252.1| DNA-damage-repair/toleration
protein-like; disease resistance protein;
polygalacturonase inhibitor-like protein [Arabidopsis
thaliana] gi|12321959|gb|AAG51016.1|AC069474_15 leucine
rich repeat protein, putative; 20015-21133 [Arabidopsis
thaliana] gi|16323097|gb|AAL15283.1| AT3g12610/T2E22_107
[Arabidopsis thaliana] gi|21592546|gb|AAM64495.1|
leucine rich repeat protein, putative [Arabidopsis
thaliana] gi|23463039|gb|AAN33189.1| At3g12610/T2E22_107
[Arabidopsis thaliana]
Length = 372
Score = 181 bits (458), Expect = 2e-44
Identities = 84/126 (66%), Positives = 99/126 (77%)
Frame = -3
Query: 874 GSVPAEFGRMRVLSTLNLDSNSLSGQIPSTLLSNSGMGILNLSRNGLEGTIPDVFGPNSY 695
G +P G M+VLS LNLD NSL+G IP +LLSNSG+ + NLSRN LEGTIPDVFG +Y
Sbjct: 245 GPIPEWMGNMKVLSLLNLDCNSLTGPIPGSLLSNSGLDVANLSRNALEGTIPDVFGSKTY 304
Query: 694 FMALDLSYNGLQGRVPGSLCSAKYIGHLDLSHNHLCGSIPMGEPFDHLEASSFSSNDCLC 515
++LDLS+N L GR+P SL SAK++GHLD+SHN LCG IP G PFDHLEA+SFS N CLC
Sbjct: 305 LVSLDLSHNSLSGRIPDSLSSAKFVGHLDISHNKLCGRIPTGFPFDHLEATSFSDNQCLC 364
Query: 514 GNPLKT 497
G PL T
Sbjct: 365 GGPLTT 370
Score = 77.8 bits (190), Expect = 2e-13
Identities = 44/118 (37%), Positives = 65/118 (54%)
Frame = -3
Query: 874 GSVPAEFGRMRVLSTLNLDSNSLSGQIPSTLLSNSGMGILNLSRNGLEGTIPDVFGPNSY 695
G +PAE G++ L+ LNL N +SG+IP++L S + L L+ NG+ G IP FG
Sbjct: 149 GEIPAEIGKLSKLAVLNLAENQMSGEIPASLTSLIELKHLELTENGITGVIPADFGSLKM 208
Query: 694 FMALDLSYNGLQGRVPGSLCSAKYIGHLDLSHNHLCGSIPMGEPFDHLEASSFSSNDC 521
+ L N L G +P S+ + + LDLS NH+ G IP E +++ S + DC
Sbjct: 209 LSRVLLGRNELTGSIPESISGMERLADLDLSKNHIEGPIP--EWMGNMKVLSLLNLDC 264
Score = 57.8 bits (138), Expect = 2e-07
Identities = 32/100 (32%), Positives = 50/100 (50%)
Frame = -3
Query: 874 GSVPAEFGRMRVLSTLNLDSNSLSGQIPSTLLSNSGMGILNLSRNGLEGTIPDVFGPNSY 695
G +P + L L+L N ++G+IP+ + S + +LNL+ N + G IP
Sbjct: 125 GEIPPCITSLASLRILDLAGNKITGEIPAEIGKLSKLAVLNLAENQMSGEIPASLTSLIE 184
Query: 694 FMALDLSYNGLQGRVPGSLCSAKYIGHLDLSHNHLCGSIP 575
L+L+ NG+ G +P S K + + L N L GSIP
Sbjct: 185 LKHLELTENGITGVIPADFGSLKMLSRVLLGRNELTGSIP 224
>emb|CAD56505.1| polygalacturonase inhibitor-like protein [Cicer arietinum]
Length = 322
Score = 179 bits (455), Expect = 4e-44
Identities = 86/127 (67%), Positives = 97/127 (75%)
Frame = -3
Query: 874 GSVPAEFGRMRVLSTLNLDSNSLSGQIPSTLLSNSGMGILNLSRNGLEGTIPDVFGPNSY 695
G +P G+M VLSTLNLD N LSG IP++L NSG+ LNLSRNGL G +PDVFG SY
Sbjct: 195 GPIPESLGKMAVLSTLNLDMNKLSGPIPASLF-NSGISDLNLSRNGLNGNLPDVFGARSY 253
Query: 694 FMALDLSYNGLQGRVPGSLCSAKYIGHLDLSHNHLCGSIPMGEPFDHLEASSFSSNDCLC 515
F LDLSYN L+G +P S+ A YIGHLDLS+NHLCG IP+G PFDHLEASSF NDCLC
Sbjct: 254 FTVLDLSYNSLKGPIPKSMGLASYIGHLDLSYNHLCGKIPVGSPFDHLEASSFVYNDCLC 313
Query: 514 GNPLKTC 494
G PLK C
Sbjct: 314 GKPLKVC 320
Score = 62.4 bits (150), Expect = 9e-09
Identities = 33/99 (33%), Positives = 53/99 (53%)
Frame = -3
Query: 871 SVPAEFGRMRVLSTLNLDSNSLSGQIPSTLLSNSGMGILNLSRNGLEGTIPDVFGPNSYF 692
++P++ GR+ L+ LN+ N++SG IP +L + + L++ N + G IP FG
Sbjct: 100 TIPSDIGRLHRLTVLNVADNAISGNIPPSLTNLRSLMHLDIRNNQISGPIPKDFGRLPML 159
Query: 691 MALDLSYNGLQGRVPGSLCSAKYIGHLDLSHNHLCGSIP 575
LS N + G +P S+ + LDLS N + G IP
Sbjct: 160 SRALLSGNKISGPIPDSISRIYRLADLDLSRNQVSGPIP 198
Score = 53.1 bits (126), Expect = 5e-06
Identities = 30/97 (30%), Positives = 50/97 (50%)
Frame = -3
Query: 865 PAEFGRMRVLSTLNLDSNSLSGQIPSTLLSNSGMGILNLSRNGLEGTIPDVFGPNSYFMA 686
PA R+ S D ++SG+IP + S + I++L N + TIP G
Sbjct: 54 PAICKLTRLSSITVADWKNISGEIPRCITSLPFLRIIDLIGNRISSTIPSDIGRLHRLTV 113
Query: 685 LDLSYNGLQGRVPGSLCSAKYIGHLDLSHNHLCGSIP 575
L+++ N + G +P SL + + + HLD+ +N + G IP
Sbjct: 114 LNVADNAISGNIPPSLTNLRSLMHLDIRNNQISGPIP 150
Score = 43.5 bits (101), Expect = 0.004
Identities = 27/100 (27%), Positives = 43/100 (43%)
Frame = -3
Query: 874 GSVPAEFGRMRVLSTLNLDSNSLSGQIPSTLLSNSGMGILNLSRNGLEGTIPDVFGPNSY 695
G +P + L ++L N +S IPS + + +LN++ N + G IP
Sbjct: 75 GEIPRCITSLPFLRIIDLIGNRISSTIPSDIGRLHRLTVLNVADNAISGNIPPSLTNLRS 134
Query: 694 FMALDLSYNGLQGRVPGSLCSAKYIGHLDLSHNHLCGSIP 575
M LD+ N + G +P + LS N + G IP
Sbjct: 135 LMHLDIRNNQISGPIPKDFGRLPMLSRALLSGNKISGPIP 174
>ref|NP_188718.1| disease resistance protein family (LRR); protein id: At3g20820.1,
supported by cDNA: gi_17380931 [Arabidopsis thaliana]
gi|9294409|dbj|BAB02490.1| polygalacturonase
inhibitor-like protein [Arabidopsis thaliana]
gi|17380932|gb|AAL36278.1| unknown protein [Arabidopsis
thaliana] gi|21436417|gb|AAM51409.1| unknown protein
[Arabidopsis thaliana]
Length = 365
Score = 177 bits (448), Expect = 2e-43
Identities = 82/127 (64%), Positives = 99/127 (77%)
Frame = -3
Query: 874 GSVPAEFGRMRVLSTLNLDSNSLSGQIPSTLLSNSGMGILNLSRNGLEGTIPDVFGPNSY 695
G++P GRM VL+TLNLD N +SG+IP TL+++S M LNLSRN L+G IP+ FGP SY
Sbjct: 237 GTIPPSLGRMSVLATLNLDGNKISGEIPQTLMTSSVMN-LNLSRNLLQGKIPEGFGPRSY 295
Query: 694 FMALDLSYNGLQGRVPGSLCSAKYIGHLDLSHNHLCGSIPMGEPFDHLEASSFSSNDCLC 515
F LDLSYN L+G +P S+ A +IGHLDLSHNHLCG IP+G PFDHLEA+SF NDCLC
Sbjct: 296 FTVLDLSYNNLKGPIPRSISGASFIGHLDLSHNHLCGRIPVGSPFDHLEAASFMFNDCLC 355
Query: 514 GNPLKTC 494
G PL+ C
Sbjct: 356 GKPLRAC 362
Score = 66.2 bits (160), Expect = 6e-10
Identities = 37/100 (37%), Positives = 54/100 (54%)
Frame = -3
Query: 874 GSVPAEFGRMRVLSTLNLDSNSLSGQIPSTLLSNSGMGILNLSRNGLEGTIPDVFGPNSY 695
G +P + GR+ L+ LN+ N +SG IP +L + S + L+L N + G IP G
Sbjct: 141 GGIPYDIGRLNRLAVLNVADNRISGSIPKSLTNLSSLMHLDLRNNLISGVIPSDVGRLKM 200
Query: 694 FMALDLSYNGLQGRVPGSLCSAKYIGHLDLSHNHLCGSIP 575
LS N + GR+P SL + + +DLS N L G+IP
Sbjct: 201 LSRALLSGNRITGRIPESLTNIYRLADVDLSGNQLYGTIP 240
Score = 55.5 bits (132), Expect = 1e-06
Identities = 33/100 (33%), Positives = 50/100 (50%)
Frame = -3
Query: 874 GSVPAEFGRMRVLSTLNLDSNSLSGQIPSTLLSNSGMGILNLSRNGLEGTIPDVFGPNSY 695
G +P R+ L TL+L N +SG IP + + + +LN++ N + G+IP S
Sbjct: 117 GEIPKCITRLPFLRTLDLIGNQISGGIPYDIGRLNRLAVLNVADNRISGSIPKSLTNLSS 176
Query: 694 FMALDLSYNGLQGRVPGSLCSAKYIGHLDLSHNHLCGSIP 575
M LDL N + G +P + K + LS N + G IP
Sbjct: 177 LMHLDLRNNLISGVIPSDVGRLKMLSRALLSGNRITGRIP 216
Score = 47.8 bits (112), Expect = 2e-04
Identities = 30/101 (29%), Positives = 50/101 (48%), Gaps = 1/101 (0%)
Frame = -3
Query: 874 GSVPAEFGRMRVLSTLNL-DSNSLSGQIPSTLLSNSGMGILNLSRNGLEGTIPDVFGPNS 698
G + A + LS + + D +SG+IP + + L+L N + G IP G +
Sbjct: 92 GHISASICELTRLSAITIADWKGISGEIPKCITRLPFLRTLDLIGNQISGGIPYDIGRLN 151
Query: 697 YFMALDLSYNGLQGRVPGSLCSAKYIGHLDLSHNHLCGSIP 575
L+++ N + G +P SL + + HLDL +N + G IP
Sbjct: 152 RLAVLNVADNRISGSIPKSLTNLSSLMHLDLRNNLISGVIP 192
>gb|AAM65656.1| leucine rich repeat protein, putative [Arabidopsis thaliana]
Length = 371
Score = 165 bits (418), Expect = 7e-40
Identities = 77/127 (60%), Positives = 97/127 (75%)
Frame = -3
Query: 874 GSVPAEFGRMRVLSTLNLDSNSLSGQIPSTLLSNSGMGILNLSRNGLEGTIPDVFGPNSY 695
G +PA FG+M VL+TLNLD N +SG IP +LL++S + LNLS N + G+IP+ FGP SY
Sbjct: 244 GPIPASFGKMSVLATLNLDGNLISGMIPGSLLASS-ISNLNLSGNLITGSIPNTFGPRSY 302
Query: 694 FMALDLSYNGLQGRVPGSLCSAKYIGHLDLSHNHLCGSIPMGEPFDHLEASSFSSNDCLC 515
F LDL+ N LQG +P S+ +A +IGHLD+SHNHLCG IPMG PFDHL+A+SF+ N CLC
Sbjct: 303 FTVLDLANNRLQGPIPASITAASFIGHLDVSHNHLCGKIPMGSPFDHLDATSFAYNACLC 362
Query: 514 GNPLKTC 494
G PL C
Sbjct: 363 GKPLGNC 369
Score = 59.3 bits (142), Expect = 7e-08
Identities = 35/100 (35%), Positives = 49/100 (49%)
Frame = -3
Query: 874 GSVPAEFGRMRVLSTLNLDSNSLSGQIPSTLLSNSGMGILNLSRNGLEGTIPDVFGPNSY 695
G +PA G++ L LNL N L G IP ++ + L+L N + G IP G
Sbjct: 148 GVIPANIGKLLRLKVLNLADNHLYGVIPPSITRLVSLSHLDLRNNNISGVIPRDIGRLKM 207
Query: 694 FMALDLSYNGLQGRVPGSLCSAKYIGHLDLSHNHLCGSIP 575
+ LS N + G++P SL + L+LS N L G IP
Sbjct: 208 VSRVLLSGNKISGQIPESLTRIYRLADLELSMNRLTGPIP 247
Score = 46.6 bits (109), Expect = 5e-04
Identities = 32/101 (31%), Positives = 50/101 (48%), Gaps = 1/101 (0%)
Frame = -3
Query: 874 GSVPAEFGRMRVLSTLNL-DSNSLSGQIPSTLLSNSGMGILNLSRNGLEGTIPDVFGPNS 698
GS+ ++ LS + + D +SG IPS + + + L+L N G IP G
Sbjct: 99 GSISPSICKLTRLSGIIIADWKGISGVIPSCIENLPFLRHLDLVGNKFSGVIPANIGKLL 158
Query: 697 YFMALDLSYNGLQGRVPGSLCSAKYIGHLDLSHNHLCGSIP 575
L+L+ N L G +P S+ + HLDL +N++ G IP
Sbjct: 159 RLKVLNLADNHLYGVIPPSITRLVSLSHLDLRNNNISGVIP 199
>gb|AAK64162.1| unknown protein [Arabidopsis thaliana]
Length = 371
Score = 165 bits (418), Expect = 7e-40
Identities = 77/127 (60%), Positives = 97/127 (75%)
Frame = -3
Query: 874 GSVPAEFGRMRVLSTLNLDSNSLSGQIPSTLLSNSGMGILNLSRNGLEGTIPDVFGPNSY 695
G +PA FG+M VL+TLNLD N +SG IP +LL++S + LNLS N + G+IP+ FGP SY
Sbjct: 244 GPIPASFGKMSVLATLNLDGNLISGMIPGSLLASS-ISNLNLSGNLITGSIPNTFGPRSY 302
Query: 694 FMALDLSYNGLQGRVPGSLCSAKYIGHLDLSHNHLCGSIPMGEPFDHLEASSFSSNDCLC 515
F LDL+ N LQG +P S+ +A +IGHLD+SHNHLCG IPMG PFDHL+A+SF+ N CLC
Sbjct: 303 FTVLDLANNRLQGLIPASITAASFIGHLDVSHNHLCGKIPMGSPFDHLDATSFAYNACLC 362
Query: 514 GNPLKTC 494
G PL C
Sbjct: 363 GKPLGNC 369
Score = 59.7 bits (143), Expect = 6e-08
Identities = 35/100 (35%), Positives = 49/100 (49%)
Frame = -3
Query: 874 GSVPAEFGRMRVLSTLNLDSNSLSGQIPSTLLSNSGMGILNLSRNGLEGTIPDVFGPNSY 695
G +PA G++ L LNL N L G IP ++ + L+L N + G IP G
Sbjct: 148 GVIPANIGKLLRLKVLNLADNHLYGVIPPSITRLVSLSHLDLRNNNISGVIPRDIGRLKM 207
Query: 694 FMALDLSYNGLQGRVPGSLCSAKYIGHLDLSHNHLCGSIP 575
+ LS N + G++P SL + L+LS N L G IP
Sbjct: 208 VSRVLLSGNKISGQIPDSLTRIYRLADLELSMNRLTGPIP 247
Score = 46.6 bits (109), Expect = 5e-04
Identities = 32/101 (31%), Positives = 50/101 (48%), Gaps = 1/101 (0%)
Frame = -3
Query: 874 GSVPAEFGRMRVLSTLNL-DSNSLSGQIPSTLLSNSGMGILNLSRNGLEGTIPDVFGPNS 698
GS+ ++ LS + + D +SG IPS + + + L+L N G IP G
Sbjct: 99 GSISPSICKLTRLSGIIIADWKGISGVIPSCIENLPFLRHLDLVGNKFSGVIPANIGKLL 158
Query: 697 YFMALDLSYNGLQGRVPGSLCSAKYIGHLDLSHNHLCGSIP 575
L+L+ N L G +P S+ + HLDL +N++ G IP
Sbjct: 159 RLKVLNLADNHLYGVIPPSITRLVSLSHLDLRNNNISGVIP 199
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 778,460,113
Number of Sequences: 1393205
Number of extensions: 18596267
Number of successful extensions: 51954
Number of sequences better than 10.0: 1271
Number of HSP's better than 10.0 without gapping: 40532
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 48119
length of database: 448,689,247
effective HSP length: 122
effective length of database: 278,718,237
effective search space used: 47103382053
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)