Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC016188A_C01 KMC016188A_c01
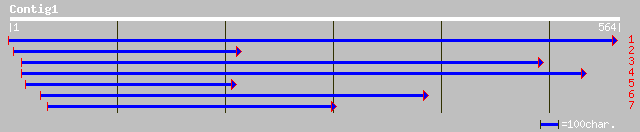
(559 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAL38267.1| unknown protein [Arabidopsis thaliana] gi|2138694... 206 1e-52
ref|NP_181440.1| unknown protein; protein id: At2g39070.1 [Arabi... 180 1e-44
dbj|BAB64743.1| P0560B06.4 [Oryza sativa (japonica cultivar-group)] 139 3e-32
ref|NP_254030.1| hypothetical protein [Pseudomonas aeruginosa PA... 49 3e-05
gb|AAO15544.1|AF458777_3 RfbB [Lactococcus lactis subsp. cremoris] 47 2e-04
>gb|AAL38267.1| unknown protein [Arabidopsis thaliana] gi|21386945|gb|AAM47876.1|
unknown protein [Arabidopsis thaliana]
Length = 351
Score = 206 bits (525), Expect = 1e-52
Identities = 97/133 (72%), Positives = 113/133 (84%)
Frame = -3
Query: 557 LLKAENVVLEFGGSVVRLSGLYKEERGAHAYWLEKGIVESRPDHVLNLIHYEDAASLSIA 378
LLKAE VVLE GG+V+RL+GLY E RGAH YWL K +++RPDH+LNLIHYEDAASL++A
Sbjct: 214 LLKAEKVVLECGGTVLRLAGLYTETRGAHTYWLSKETIDARPDHILNLIHYEDAASLAVA 273
Query: 377 ILKKQFRGRIFLGCDNHPLSRQEVMDLVNKSGKFSKKFEKFTGSDDPLGKRLNNSKTRQE 198
I+KK+ RIFLGCDNHPLSRQEVMDL+ +SGKF KKF+ FT + PLGK+LNNSKTR E
Sbjct: 274 IMKKKAGARIFLGCDNHPLSRQEVMDLMAQSGKFDKKFKGFTSTSGPLGKKLNNSKTRAE 333
Query: 197 IGWEPKYSSFAHF 159
IGWEPKY SFA F
Sbjct: 334 IGWEPKYPSFAQF 346
>ref|NP_181440.1| unknown protein; protein id: At2g39070.1 [Arabidopsis thaliana]
gi|25370805|pir||G84812 hypothetical protein At2g39070
[imported] - Arabidopsis thaliana
gi|3928091|gb|AAC79617.1| unknown protein [Arabidopsis
thaliana]
Length = 124
Score = 180 bits (456), Expect = 1e-44
Identities = 83/116 (71%), Positives = 97/116 (83%)
Frame = -3
Query: 506 LSGLYKEERGAHAYWLEKGIVESRPDHVLNLIHYEDAASLSIAILKKQFRGRIFLGCDNH 327
L+ LY E RGAH YWL K +++RPDH+LNLIHYEDAASL++AI+KK+ RIFLGCDNH
Sbjct: 4 LNFLYTETRGAHTYWLSKETIDARPDHILNLIHYEDAASLAVAIMKKKAGARIFLGCDNH 63
Query: 326 PLSRQEVMDLVNKSGKFSKKFEKFTGSDDPLGKRLNNSKTRQEIGWEPKYSSFAHF 159
PLSRQEVMDL+ +SGKF KKF+ FT + PLGK+LNNSKTR EIGWEPKY SFA F
Sbjct: 64 PLSRQEVMDLMAQSGKFDKKFKGFTSTSGPLGKKLNNSKTRAEIGWEPKYPSFAQF 119
>dbj|BAB64743.1| P0560B06.4 [Oryza sativa (japonica cultivar-group)]
Length = 151
Score = 139 bits (349), Expect = 3e-32
Identities = 73/134 (54%), Positives = 86/134 (63%)
Frame = -3
Query: 557 LLKAENVVLEFGGSVVRLSGLYKEERGAHAYWLEKGIVESRPDHVLNLIHYEDAASLSIA 378
LLKAEN VLE GGSV+RL DAASL+IA
Sbjct: 48 LLKAENAVLEAGGSVLRL----------------------------------DAASLAIA 73
Query: 377 ILKKQFRGRIFLGCDNHPLSRQEVMDLVNKSGKFSKKFEKFTGSDDPLGKRLNNSKTRQE 198
I+K++ R R+F+GCDN PLSRQE+MDLVN+SGKF KF+ FTG+D PLGKR+ NSKTR E
Sbjct: 74 IMKRRLRARVFVGCDNQPLSRQEIMDLVNRSGKFDTKFQGFTGTDGPLGKRMENSKTRAE 133
Query: 197 IGWEPKYSSFAHFL 156
IGW+PKY SF FL
Sbjct: 134 IGWQPKYPSFTEFL 147
>ref|NP_254030.1| hypothetical protein [Pseudomonas aeruginosa PA01]
gi|11350423|pir||E82979 hypothetical protein PA5343
[imported] - Pseudomonas aeruginosa (strain PAO1)
gi|9951662|gb|AAG08728.1|AE004946_12 hypothetical
protein [Pseudomonas aeruginosa PAO1]
Length = 283
Score = 49.3 bits (116), Expect = 3e-05
Identities = 43/141 (30%), Positives = 71/141 (49%), Gaps = 11/141 (7%)
Frame = -3
Query: 557 LLKAENVVLEFG--GSVVRLSGLYKEERGAHAYWLEKGI-VESRPDHVLNLIHYEDAASL 387
+L AE V L+ G + VRL+G+Y R + +G V S P N IH +DAA L
Sbjct: 138 MLDAEQVALDSGIPATRVRLAGIYGPGREWLLNQVRQGYRVVSEPPLYANRIHADDAAGL 197
Query: 386 SIAILKKQFRGRI----FLGCDNHPLSRQEVMDLVNKSGKFSKKFEKFT----GSDDPLG 231
+L+ G+ ++G D+ P++ EV+D + + S+ ++ + GS
Sbjct: 198 LAFLLRADAGGQALEDCYIGVDDAPVAMHEVVDYLRQRLGVSQWADEHSVRRAGS----- 252
Query: 230 KRLNNSKTRQEIGWEPKYSSF 168
KR +N + R +GW P+Y S+
Sbjct: 253 KRCSNRRARA-LGWVPRYPSY 272
>gb|AAO15544.1|AF458777_3 RfbB [Lactococcus lactis subsp. cremoris]
Length = 350
Score = 46.6 bits (109), Expect = 2e-04
Identities = 28/89 (31%), Positives = 48/89 (53%), Gaps = 2/89 (2%)
Frame = -3
Query: 428 HVLNLIHYEDAASLSIAILKKQFRGRIFL-GCDNHPLSRQEVMDLVNKSGKFSKKFEKFT 252
+V + IH ED +S IL K G +L G D +++ + D++ + GK +++ T
Sbjct: 224 NVRDWIHTEDHSSGVWTILTKGQMGETYLIGADGEKNNKEVLEDILTRMGKDKDDYDRVT 283
Query: 251 G-SDDPLGKRLNNSKTRQEIGWEPKYSSF 168
+ L ++NSK R E+GW PK++ F
Sbjct: 284 DRAGHDLRYAIDNSKLRTELGWTPKHTDF 312
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 494,132,256
Number of Sequences: 1393205
Number of extensions: 10383189
Number of successful extensions: 23397
Number of sequences better than 10.0: 70
Number of HSP's better than 10.0 without gapping: 22879
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 23394
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19808345223
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)