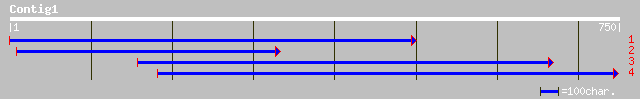
Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC016087A_C01 KMC016087A_c01
(742 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM98162.1| unknown protein [Arabidopsis thaliana] 116 4e-25
ref|NP_683492.1| hypothetical protein; protein id: At1g73885.1 [... 112 6e-24
gb|AAK38498.1|AC087181_14 unknown protein [Oryza sativa] 111 9e-24
ref|NP_486662.1| ATP-binding protein of ABC transporter [Nostoc ... 35 1.1
emb|CAD37158.1| hypothetical protein [Aspergillus fumigatus] 33 5.5
>gb|AAM98162.1| unknown protein [Arabidopsis thaliana]
Length = 212
Score = 116 bits (290), Expect = 4e-25
Identities = 53/90 (58%), Positives = 72/90 (79%), Gaps = 1/90 (1%)
Frame = -3
Query: 650 LPGSEVDFFEGPQWDTVGLMGEYLWVAGVVFAILAGLFGAANYNQGASDFKDTPAYKESV 471
LPG E D FEG +WD +G +YLW G++FA+++G A YN+GA+DFK+TP YKE++
Sbjct: 123 LPGLEPDPFEGEKWDGLGFFVQYLWAFGILFALISGGLAAGTYNEGATDFKETPVYKEAI 182
Query: 470 QSRELLEQPDASDS-DVFESNPTEVAPSLE 384
QSR+LL++ ++S+S DVFESNPTEVAPSLE
Sbjct: 183 QSRDLLDEAESSNSEDVFESNPTEVAPSLE 212
>ref|NP_683492.1| hypothetical protein; protein id: At1g73885.1 [Arabidopsis
thaliana]
Length = 192
Score = 112 bits (280), Expect = 6e-24
Identities = 50/89 (56%), Positives = 71/89 (79%), Gaps = 1/89 (1%)
Frame = -3
Query: 650 LPGSEVDFFEGPQWDTVGLMGEYLWVAGVVFAILAGLFGAANYNQGASDFKDTPAYKESV 471
LPG E D FEG +WD +G +YLW G++FA+++G A YN+GA+DFK+TP YKE++
Sbjct: 91 LPGLEPDPFEGEKWDGLGFFVQYLWAFGILFALISGGLAAGTYNEGATDFKETPVYKEAI 150
Query: 470 QSRELLEQPDASDS-DVFESNPTEVAPSL 387
QSR+LL++ ++S+S DVFESNPTEVAP++
Sbjct: 151 QSRDLLDEAESSNSEDVFESNPTEVAPTI 179
>gb|AAK38498.1|AC087181_14 unknown protein [Oryza sativa]
Length = 199
Score = 111 bits (278), Expect = 9e-24
Identities = 57/125 (45%), Positives = 77/125 (61%), Gaps = 12/125 (9%)
Frame = -3
Query: 722 ADQEPQWTAEIGAGTGND------GYPDRALPGSEV------DFFEGPQWDTVGLMGEYL 579
A P + AG G D G+ P E+ DF+EGPQWD +G +Y+
Sbjct: 75 APAPPDMDEDAAAGEGVDWEGEPLGFEVSTTPMPELPDPEKPDFWEGPQWDALGFFVQYM 134
Query: 578 WVAGVVFAILAGLFGAANYNQGASDFKDTPAYKESVQSRELLEQPDASDSDVFESNPTEV 399
W GV F ++A F A YN+GA+DF++TP+YKESVQ++E E+ ++S SDVFE NPTEV
Sbjct: 135 WAFGVFFGLVACGFAVATYNEGATDFRETPSYKESVQTQEFPEESESSGSDVFEGNPTEV 194
Query: 398 APSLE 384
AP+LE
Sbjct: 195 APALE 199
>ref|NP_486662.1| ATP-binding protein of ABC transporter [Nostoc sp. PCC 7120]
gi|25307498|pir||AG2133 ATP-binding protein of ABC
transporter all2622 [imported] - Nostoc sp. (strain PCC
7120) gi|17131715|dbj|BAB74321.1| ATP-binding protein of
ABC transporter [Nostoc sp. PCC 7120]
Length = 573
Score = 35.0 bits (79), Expect = 1.1
Identities = 23/62 (37%), Positives = 32/62 (51%)
Frame = -1
Query: 628 SLKGPSGIQLA*WASICGLLVLFLRYLLACLVPQIIIKEHQILRILLHTRNLFSLENF*N 449
SL G + L+ WA I G + L L ++L L+ +I +H+ L RNL LE N
Sbjct: 17 SLTGKAQFGLSPWAFIAGFVALNLGHILVILIGRITKTQHRFTMRSLLRRNL--LERLLN 74
Query: 448 NP 443
NP
Sbjct: 75 NP 76
>emb|CAD37158.1| hypothetical protein [Aspergillus fumigatus]
Length = 525
Score = 32.7 bits (73), Expect = 5.5
Identities = 21/60 (35%), Positives = 28/60 (46%)
Frame = +2
Query: 530 RHQTSQQVSQKQHQQPTDTRPSSQLYPTGALQRNPLHCPEAPDRGTHHSLCRRQSPPSTA 709
+ +SQ SQKQ Q P+D +P S P LQ P P+ P G + + PP A
Sbjct: 349 QRSSSQPNSQKQVQPPSDPQPRSHPSPQVQLQAQPPSQPQ-PKPGPSIQIRTKTQPPPKA 407
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 659,307,502
Number of Sequences: 1393205
Number of extensions: 14948169
Number of successful extensions: 44813
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 42300
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 44695
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 35469585522
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)