Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC016073A_C01 KMC016073A_c01
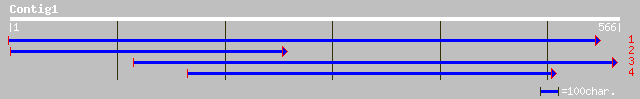
(565 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_188902.1| 20S proteasome beta subunit D (PBD1); protein i... 144 6e-34
ref|NP_193216.1| 20S proteasome beta subunit D2 (PBD2); protein ... 137 1e-31
gb|AAO19369.1| 20S proteasome beta 4 subunit [Oryza sativa (japo... 120 1e-26
sp|Q9LST6|PSB2_ORYSA Proteasome subunit beta type 2 (20S proteas... 120 1e-26
dbj|BAC37303.1| unnamed protein product [Mus musculus] 63 3e-09
>ref|NP_188902.1| 20S proteasome beta subunit D (PBD1); protein id: At3g22630.1,
supported by cDNA: gi_15529249, supported by cDNA:
gi_3421110 [Arabidopsis thaliana]
gi|17380179|sp|O23714|PS21_ARATH Proteasome subunit beta
type 2-1 (20S proteasome alpha subunit D1)
gi|25330487|pir||T51982 C 3.4.25.1 proteasome
endopeptidase complex () beta chain PBD1 [imported] -
Arabidopsis thaliana gi|2511590|emb|CAA74026.1|
multicatalytic endopeptidase complex, proteasome
component, beta subunit [Arabidopsis thaliana]
gi|3421111|gb|AAC32070.1| 20S proteasome beta subunit
PBD1 [Arabidopsis thaliana] gi|11994294|dbj|BAB01477.1|
multicatalytic endopeptidase complex, proteasome
component, beta subunit [Arabidopsis thaliana]
gi|15529250|gb|AAK97719.1| AT3g22630/F16J14_20
[Arabidopsis thaliana] gi|23505793|gb|AAN28756.1|
At3g22630/F16J14_20 [Arabidopsis thaliana]
Length = 204
Score = 144 bits (364), Expect = 6e-34
Identities = 70/78 (89%), Positives = 74/78 (94%)
Frame = -3
Query: 563 ATLHKLEKGAFGYGSYFSLSMMDRHYHSGMSVEEAIDLVDKCIHEIRSRLVVAPPNFVIK 384
ATLHK++KGAFGYGSYFSLS MDRHY S MSVEEAI+LVDKCI EIRSRLVVAPPNFVIK
Sbjct: 121 ATLHKVDKGAFGYGSYFSLSTMDRHYRSDMSVEEAIELVDKCILEIRSRLVVAPPNFVIK 180
Query: 383 IVDKDGAREYAWRQSVKD 330
IVDKDGAR+YAWRQSVKD
Sbjct: 181 IVDKDGARDYAWRQSVKD 198
>ref|NP_193216.1| 20S proteasome beta subunit D2 (PBD2); protein id: At4g14800.1,
supported by cDNA: 24266., supported by cDNA: gi_3421113
[Arabidopsis thaliana] gi|17380180|sp|O24633|PS22_ARATH
Proteasome subunit beta type 2-2 (20S proteasome alpha
subunit D2) gi|7451296|pir||A71411 C 3.4.25.1 proteasome
endopeptidase complex () chain PBD2 [imported] -
Arabidopsis thaliana gi|2244837|emb|CAB10259.1|
proteasome chain protein [Arabidopsis thaliana]
gi|2511572|emb|CAA73618.1| multicatalytic endopeptidase
[Arabidopsis thaliana] gi|3421114|gb|AAC32071.1| 20S
proteasome beta subunit PBD2 [Arabidopsis thaliana]
gi|7268226|emb|CAB78522.1| proteasome chain protein
[Arabidopsis thaliana] gi|21592467|gb|AAM64418.1|
proteasome chain protein [Arabidopsis thaliana]
Length = 199
Score = 137 bits (344), Expect = 1e-31
Identities = 66/78 (84%), Positives = 70/78 (89%)
Frame = -3
Query: 563 ATLHKLEKGAFGYGSYFSLSMMDRHYHSGMSVEEAIDLVDKCIHEIRSRLVVAPPNFVIK 384
ATLHK++KGAFGYGSYFSLS MDRHY S MSVEEAI+LVDKCI EIRSRLV+APPNFVIK
Sbjct: 121 ATLHKVDKGAFGYGSYFSLSTMDRHYRSDMSVEEAIELVDKCILEIRSRLVIAPPNFVIK 180
Query: 383 IVDKDGAREYAWRQSVKD 330
IVDKDGAREY WR S D
Sbjct: 181 IVDKDGAREYGWRISTAD 198
>gb|AAO19369.1| 20S proteasome beta 4 subunit [Oryza sativa (japonica
cultivar-group)]
Length = 212
Score = 120 bits (301), Expect = 1e-26
Identities = 60/83 (72%), Positives = 69/83 (82%)
Frame = -3
Query: 563 ATLHKLEKGAFGYGSYFSLSMMDRHYHSGMSVEEAIDLVDKCIHEIRSRLVVAPPNFVIK 384
AT HK+EKGAFGYGSYF LS+MD+ Y MSVEEA+DLVDKCI EIR RLVVAP NF+IK
Sbjct: 121 ATFHKIEKGAFGYGSYFCLSLMDKLYRPDMSVEEAVDLVDKCIKEIRLRLVVAPQNFIIK 180
Query: 383 IVDKDGAREYAWRQSVKDTPAPA 315
IVDK+GAREYA R++ D+P A
Sbjct: 181 IVDKEGAREYA-RRAYTDSPPEA 202
>sp|Q9LST6|PSB2_ORYSA Proteasome subunit beta type 2 (20S proteasome alpha subunit D)
(20S proteasome subunit beta-4)
gi|8671508|dbj|BAA96837.1| beta 4 subunit of 20S
proteasome [Oryza sativa (japonica cultivar-group)]
Length = 212
Score = 120 bits (301), Expect = 1e-26
Identities = 60/83 (72%), Positives = 69/83 (82%)
Frame = -3
Query: 563 ATLHKLEKGAFGYGSYFSLSMMDRHYHSGMSVEEAIDLVDKCIHEIRSRLVVAPPNFVIK 384
AT HK+EKGAFGYGSYF LS+MD+ Y MSVEEA+DLVDKCI EIR RLVVAP NF+IK
Sbjct: 121 ATFHKIEKGAFGYGSYFCLSLMDKLYRPDMSVEEAVDLVDKCIKEIRLRLVVAPQNFIIK 180
Query: 383 IVDKDGAREYAWRQSVKDTPAPA 315
IVDK+GAREYA R++ D+P A
Sbjct: 181 IVDKEGAREYA-RRAYTDSPPEA 202
>dbj|BAC37303.1| unnamed protein product [Mus musculus]
Length = 201
Score = 62.8 bits (151), Expect = 3e-09
Identities = 26/66 (39%), Positives = 45/66 (67%)
Frame = -3
Query: 563 ATLHKLEKGAFGYGSYFSLSMMDRHYHSGMSVEEAIDLVDKCIHEIRSRLVVAPPNFVIK 384
A L K A GYG++ +LS++DR+Y +S E A++L+ KC+ E++ R ++ P F ++
Sbjct: 122 AALAKAPFAAHGYGAFLTLSILDRYYTPTISRERAVELLRKCLEELQKRFILNLPTFSVR 181
Query: 383 IVDKDG 366
++DKDG
Sbjct: 182 VIDKDG 187
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 497,042,790
Number of Sequences: 1393205
Number of extensions: 10557133
Number of successful extensions: 40821
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 38060
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 40576
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20382500157
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)