Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC016072A_C01 KMC016072A_c01
(528 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
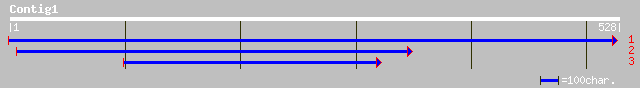
ref|NP_188394.1| hypothetical protein; protein id: At3g17670.1 [... 127 8e-29
dbj|BAB02058.1| gb|AAF19538.1~gene_id:MKP6.24~similar to unknown... 127 8e-29
ref|NP_061945.1| hypothetical protein LOC54557 [Homo sapiens] gi... 60 2e-08
gb|AAM98143.1| stress-induced protein sti1-like protein [Arabido... 58 6e-08
ref|NP_192977.1| stress-induced protein sti1 -like protein; prot... 58 6e-08
>ref|NP_188394.1| hypothetical protein; protein id: At3g17670.1 [Arabidopsis
thaliana]
Length = 366
Score = 127 bits (319), Expect = 8e-29
Identities = 62/117 (52%), Positives = 87/117 (73%)
Frame = -1
Query: 516 EFEKRNFVEAELLLSQAIDLKSFGGMHVTFKCRSLVRLELGNYSGALNDATEALALAPRY 337
+ + NF EAE LL+QA++LK +GG+H FK RS+ +L + +YSGAL D ++ALALAP Y
Sbjct: 250 QIDAGNFSEAEALLTQALELKPYGGLHRIFKHRSVAKLGMLDYSGALEDISQALALAPNY 309
Query: 336 SEAYICVGNAFLALDKFDLAEQSYLAALDIDPSIRHSKSFKVRIAMLKEKLAAANRT 166
SE YIC G+ ++A ++DLAE+SYL L+IDPS+R SK FK RIA L++K+ + T
Sbjct: 310 SEPYICQGDVYVAKGQYDLAEKSYLTCLEIDPSLRRSKPFKARIAKLQQKVVELDVT 366
>dbj|BAB02058.1| gb|AAF19538.1~gene_id:MKP6.24~similar to unknown protein
[Arabidopsis thaliana]
Length = 236
Score = 127 bits (319), Expect = 8e-29
Identities = 62/117 (52%), Positives = 87/117 (73%)
Frame = -1
Query: 516 EFEKRNFVEAELLLSQAIDLKSFGGMHVTFKCRSLVRLELGNYSGALNDATEALALAPRY 337
+ + NF EAE LL+QA++LK +GG+H FK RS+ +L + +YSGAL D ++ALALAP Y
Sbjct: 120 QIDAGNFSEAEALLTQALELKPYGGLHRIFKHRSVAKLGMLDYSGALEDISQALALAPNY 179
Query: 336 SEAYICVGNAFLALDKFDLAEQSYLAALDIDPSIRHSKSFKVRIAMLKEKLAAANRT 166
SE YIC G+ ++A ++DLAE+SYL L+IDPS+R SK FK RIA L++K+ + T
Sbjct: 180 SEPYICQGDVYVAKGQYDLAEKSYLTCLEIDPSLRRSKPFKARIAKLQQKVVELDVT 236
>ref|NP_061945.1| hypothetical protein LOC54557 [Homo sapiens]
gi|15082283|gb|AAH12044.1|AAH12044 Similar to small
glutamine-rich tetratricopeptide repeat (TPR)-containing
[Homo sapiens] gi|21755789|dbj|BAC04761.1| unnamed
protein product [Homo sapiens]
Length = 304
Score = 59.7 bits (143), Expect = 2e-08
Identities = 36/109 (33%), Positives = 65/109 (59%)
Frame = -1
Query: 510 EKRNFVEAELLLSQAIDLKSFGGMHVTFKCRSLVRLELGNYSGALNDATEALALAPRYSE 331
++ N+ A +QAI+L ++ + R+ + +LG+Y+ A+ D +A+A+ +YS+
Sbjct: 97 KEENYAAAVDCYTQAIELDPNNAVY--YCNRAAAQSKLGHYTDAIKDCEKAIAIDSKYSK 154
Query: 330 AYICVGNAFLALDKFDLAEQSYLAALDIDPSIRHSKSFKVRIAMLKEKL 184
AY +G A AL+KF+ A SY ALD+DP + S+K + + ++KL
Sbjct: 155 AYGRMGLALTALNKFEEAVTSYQKALDLDP---ENDSYKSNLKIAEQKL 200
>gb|AAM98143.1| stress-induced protein sti1-like protein [Arabidopsis thaliana]
Length = 530
Score = 58.2 bits (139), Expect = 6e-08
Identities = 40/121 (33%), Positives = 59/121 (48%)
Frame = -1
Query: 528 RADIEFEKRNFVEAELLLSQAIDLKSFGGMHVTFKCRSLVRLELGNYSGALNDATEALAL 349
+ + F ++ A ++AI+L H+ + RS L Y AL+DA + + L
Sbjct: 8 KGNAAFSSGDYATAITHFTEAINLSPTN--HILYSNRSASYASLHRYEEALSDAKKTIEL 65
Query: 348 APRYSEAYICVGNAFLALDKFDLAEQSYLAALDIDPSIRHSKSFKVRIAMLKEKLAAANR 169
P +S+ Y +G AF+ L KFD A SY L+IDPS MLK LA A+R
Sbjct: 66 KPDWSKGYSRLGAAFIGLSKFDEAVDSYKKGLEIDPSNE----------MLKSGLADASR 115
Query: 168 T 166
+
Sbjct: 116 S 116
>ref|NP_192977.1| stress-induced protein sti1 -like protein; protein id: At4g12400.1
[Arabidopsis thaliana] gi|11346440|pir||T48150
stress-induced protein sti1-like protein - Arabidopsis
thaliana gi|5281051|emb|CAB45987.1| stress-induced
protein sti1-like protein [Arabidopsis thaliana]
gi|7267942|emb|CAB78283.1| stress-induced protein
sti1-like protein [Arabidopsis thaliana]
Length = 558
Score = 58.2 bits (139), Expect = 6e-08
Identities = 40/121 (33%), Positives = 59/121 (48%)
Frame = -1
Query: 528 RADIEFEKRNFVEAELLLSQAIDLKSFGGMHVTFKCRSLVRLELGNYSGALNDATEALAL 349
+ + F ++ A ++AI+L H+ + RS L Y AL+DA + + L
Sbjct: 8 KGNAAFSSGDYATAITHFTEAINLSPTN--HILYSNRSASYASLHRYEEALSDAKKTIEL 65
Query: 348 APRYSEAYICVGNAFLALDKFDLAEQSYLAALDIDPSIRHSKSFKVRIAMLKEKLAAANR 169
P +S+ Y +G AF+ L KFD A SY L+IDPS MLK LA A+R
Sbjct: 66 KPDWSKGYSRLGAAFIGLSKFDEAVDSYKKGLEIDPSNE----------MLKSGLADASR 115
Query: 168 T 166
+
Sbjct: 116 S 116
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 402,352,220
Number of Sequences: 1393205
Number of extensions: 8017123
Number of successful extensions: 21624
Number of sequences better than 10.0: 382
Number of HSP's better than 10.0 without gapping: 20681
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 21589
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 17308240320
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)