Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
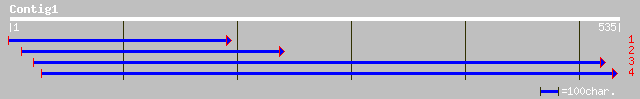
Query= KMC015974A_C01 KMC015974A_c01
(534 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_178345.1| putative cinnamoyl-CoA reductase; protein id: A... 161 4e-39
dbj|BAC57643.1| putative cinnamoyl-CoA reductase [Oryza sativa (... 123 1e-27
gb|AAO60009.1| putative cinnamoyl-CoA reductase [Oryza sativa (j... 114 7e-25
ref|NP_200657.1| cinnamoyl-CoA reductase - like protein; protein... 107 1e-22
gb|AAG13987.1|AF298828_1 putative cinnamoyl-CoA reductase [Prunu... 103 2e-21
>ref|NP_178345.1| putative cinnamoyl-CoA reductase; protein id: At2g02400.1
[Arabidopsis thaliana] gi|25284019|pir||C84436 probable
cinnamoyl-CoA reductase [imported] - Arabidopsis
thaliana gi|3894172|gb|AAC78522.1| putative
cinnamoyl-CoA reductase [Arabidopsis thaliana]
gi|28973736|gb|AAO64184.1| putative cinnamoyl-CoA
reductase [Arabidopsis thaliana]
Length = 318
Score = 161 bits (408), Expect = 4e-39
Identities = 73/104 (70%), Positives = 91/104 (87%)
Frame = -3
Query: 532 QEHYWLGAVHVKDVARAQVLLFETPTAAGRYLCTNGIYQFSSFASIVSELYPDFPVHRFP 353
QEH+WLG VHVKDVA+ V+LFETP A+GR+LCTNGIYQFS FA++VS+L+P+F VH+F
Sbjct: 215 QEHHWLGVVHVKDVAKGHVMLFETPDASGRFLCTNGIYQFSEFAALVSKLFPEFAVHKFD 274
Query: 352 EETQPGLTPCKDAAKRLIDLGLVFTPIQDAVRDAVESLIAKGFL 221
+ETQPGLT C DAAKRLI+LGLVFT ++DAV++ V+SL KGFL
Sbjct: 275 KETQPGLTSCNDAAKRLIELGLVFTAVEDAVKETVQSLRDKGFL 318
>dbj|BAC57643.1| putative cinnamoyl-CoA reductase [Oryza sativa (japonica
cultivar-group)]
Length = 327
Score = 123 bits (309), Expect = 1e-27
Identities = 63/109 (57%), Positives = 80/109 (72%), Gaps = 5/109 (4%)
Frame = -3
Query: 532 QEHYWLGAVHVKDVARAQVLLFETPTAAGRYLCTNGIYQFSSFASIVSELYPDF--PVHR 359
Q YWLGAVHV+DVA A +LL E PT +GRYLCTNGIYQFS FA + + + P + +HR
Sbjct: 218 QADYWLGAVHVRDVAAAHLLLLEAPTVSGRYLCTNGIYQFSDFARLAARICPAYAHAIHR 277
Query: 358 FPE-ETQPGLTP--CKDAAKRLIDLGLVFTPIQDAVRDAVESLIAKGFL 221
F E TQP L P +DAA+RL+DLGLV TP+++A++DA +SL K FL
Sbjct: 278 FEEGTTQPWLVPRDARDAARRLLDLGLVLTPLEEAIKDAEKSLTDKCFL 326
>gb|AAO60009.1| putative cinnamoyl-CoA reductase [Oryza sativa (japonica
cultivar-group)] gi|29124112|gb|AAO65853.1| putative
cinnamoyl-CoA reductase [Oryza sativa (japonica
cultivar-group)]
Length = 334
Score = 114 bits (285), Expect = 7e-25
Identities = 52/101 (51%), Positives = 78/101 (76%)
Frame = -3
Query: 523 YWLGAVHVKDVARAQVLLFETPTAAGRYLCTNGIYQFSSFASIVSELYPDFPVHRFPEET 344
+++G VHV+DVA A +LL+E P+A+GR+LC I +S FAS V+ELYP++ V + P+ET
Sbjct: 233 FYMGPVHVEDVALAHILLYENPSASGRHLCVQSIAHWSDFASKVAELYPEYKVPKLPKET 292
Query: 343 QPGLTPCKDAAKRLIDLGLVFTPIQDAVRDAVESLIAKGFL 221
QPGL + A+K+LI LGL F+P++ +RD+VESL ++GF+
Sbjct: 293 QPGLVRAEAASKKLIALGLQFSPMEKIIRDSVESLKSRGFI 333
>ref|NP_200657.1| cinnamoyl-CoA reductase - like protein; protein id: At5g58490.1,
supported by cDNA: 30064., supported by cDNA:
gi_17979314 [Arabidopsis thaliana]
gi|10177026|dbj|BAB10264.1| dihydroflavonol
4-reductase-like [Arabidopsis thaliana]
gi|21592589|gb|AAM64538.1| cinnamoyl-CoA reductase-like
protein [Arabidopsis thaliana]
gi|27754235|gb|AAO22571.1| putative cinnamoyl-CoA
reductase [Arabidopsis thaliana]
Length = 324
Score = 107 bits (266), Expect = 1e-22
Identities = 48/103 (46%), Positives = 75/103 (72%)
Frame = -3
Query: 529 EHYWLGAVHVKDVARAQVLLFETPTAAGRYLCTNGIYQFSSFASIVSELYPDFPVHRFPE 350
E++++G+VH KDVA A +L++E P + GR+LC I + F + V+ELYP++ V + P
Sbjct: 221 ENFFMGSVHFKDVALAHILVYEDPYSKGRHLCVEAISHYGDFVAKVAELYPNYNVPKLPR 280
Query: 349 ETQPGLTPCKDAAKRLIDLGLVFTPIQDAVRDAVESLIAKGFL 221
ETQPGL K+A+K+LIDLGL F +++ +++ VESL +KGF+
Sbjct: 281 ETQPGLLRDKNASKKLIDLGLKFISMEEIIKEGVESLKSKGFI 323
>gb|AAG13987.1|AF298828_1 putative cinnamoyl-CoA reductase [Prunus avium]
Length = 159
Score = 103 bits (256), Expect = 2e-21
Identities = 49/103 (47%), Positives = 71/103 (68%)
Frame = -3
Query: 529 EHYWLGAVHVKDVARAQVLLFETPTAAGRYLCTNGIYQFSSFASIVSELYPDFPVHRFPE 350
E+ ++G+VH KDVA A +LL E +A GR+LC I + F + V+ELYP++ V P+
Sbjct: 56 ENIFMGSVHFKDVALAHILLHENKSATGRHLCVEAISHYGDFVAKVAELYPEYKVPSLPK 115
Query: 349 ETQPGLTPCKDAAKRLIDLGLVFTPIQDAVRDAVESLIAKGFL 221
+TQPGL K+ AK+L++LGL F P+ ++DAVESL KGF+
Sbjct: 116 DTQPGLLREKNGAKKLMNLGLDFIPMDQIIKDAVESLKNKGFI 158
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 446,430,811
Number of Sequences: 1393205
Number of extensions: 9133077
Number of successful extensions: 17975
Number of sequences better than 10.0: 162
Number of HSP's better than 10.0 without gapping: 17461
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 17918
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 17885181664
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)