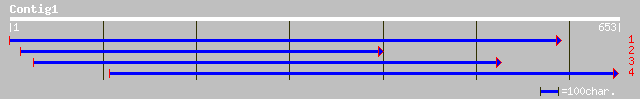
Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC015932A_C01 KMC015932A_c01
(653 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAB56195.1| Knolle [Capsicum annuum] 230 1e-59
emb|CAD78064.1| knolle [Antirrhinum majus] 222 4e-57
ref|NP_172332.1| syntaxin SYP111 (KNOLLE); protein id: At1g08560... 207 8e-53
ref|NP_187788.1| syntaxin SYP121; protein id: At3g11820.1, suppo... 169 2e-41
ref|NP_172591.1| syntaxin SYP125; protein id: At1g11250.1 [Arabi... 161 8e-39
>emb|CAB56195.1| Knolle [Capsicum annuum]
Length = 311
Score = 230 bits (587), Expect = 1e-59
Identities = 113/145 (77%), Positives = 130/145 (88%), Gaps = 1/145 (0%)
Frame = -1
Query: 653 RYFTVTGENPDEEVIEKIISNGD-EGGFMFLGKPVEEHGRGKVLETVAEIQDRHEGAKEV 477
RYFTVTGE PDEEVI+KIIS+G+ +GG FL + ++EHGRGKVLETV EIQDRH+ AKE+
Sbjct: 167 RYFTVTGEQPDEEVIDKIISSGNGQGGEEFLSRAIQEHGRGKVLETVVEIQDRHDAAKEI 226
Query: 476 EKSLLELHQVFLDMAVMVEAQGEKMDDIEHHVLHSSHYVKDGTKNLQTAKMYQKSSRKWM 297
EKSLLELHQ+FLDMAVMVEAQGEKMDDIEHHV++++ YV DGTKNL+TAK YQKSSR WM
Sbjct: 227 EKSLLELHQIFLDMAVMVEAQGEKMDDIEHHVVNAAQYVNDGTKNLKTAKEYQKSSRTWM 286
Query: 296 CIGIIILLILILVIVIPIVTSLTSS 222
CIGIIILLILILV++IPI TS T S
Sbjct: 287 CIGIIILLILILVVIIPIATSFTKS 311
>emb|CAD78064.1| knolle [Antirrhinum majus]
Length = 307
Score = 222 bits (565), Expect = 4e-57
Identities = 108/144 (75%), Positives = 128/144 (88%)
Frame = -1
Query: 653 RYFTVTGENPDEEVIEKIISNGDEGGFMFLGKPVEEHGRGKVLETVAEIQDRHEGAKEVE 474
RY+TVTGENPDEEVI+KIISNG E FL + ++EHGRG+VLETV EIQDRH+ AKE+E
Sbjct: 167 RYYTVTGENPDEEVIDKIISNGGEE---FLSRAIQEHGRGRVLETVVEIQDRHDAAKEIE 223
Query: 473 KSLLELHQVFLDMAVMVEAQGEKMDDIEHHVLHSSHYVKDGTKNLQTAKMYQKSSRKWMC 294
KSLLELHQ+FLDMAVMVEAQGEKMDDIEHHV++++ YV DGTKNL+TAK YQKSSR+ MC
Sbjct: 224 KSLLELHQIFLDMAVMVEAQGEKMDDIEHHVMNAAQYVSDGTKNLKTAKDYQKSSRRCMC 283
Query: 293 IGIIILLILILVIVIPIVTSLTSS 222
IGII+LL+LILV++IPI TS + S
Sbjct: 284 IGIILLLVLILVVIIPIATSFSKS 307
>ref|NP_172332.1| syntaxin SYP111 (KNOLLE); protein id: At1g08560.1, supported by
cDNA: gi_1184164, supported by cDNA: gi_18377780
[Arabidopsis thaliana] gi|2501102|sp|Q42374|S111_ARATH
Syntaxin-related protein KNOLLE (Syntaxin 111)
(AtSYP111) gi|7488363|pir||T00709 syntaxin-related
protein homolog F22O13.4 - Arabidopsis thaliana
gi|1184165|gb|AAC49162.1| syntaxin-related
gi|1184167|gb|AAC49163.1| syntaxin-related
gi|24030428|gb|AAN41370.1| putative syntaxin-related
protein [Arabidopsis thaliana] gi|1587182|prf||2206310A
syntaxin-related protein
Length = 310
Score = 207 bits (528), Expect = 8e-53
Identities = 98/144 (68%), Positives = 129/144 (89%)
Frame = -1
Query: 653 RYFTVTGENPDEEVIEKIISNGDEGGFMFLGKPVEEHGRGKVLETVAEIQDRHEGAKEVE 474
RYFTVTGE+ ++E+IEKII++ + GG FL + ++EHG+GKVLETV EIQDR++ AKE+E
Sbjct: 168 RYFTVTGEHANDEMIEKIITD-NAGGEEFLTRAIQEHGKGKVLETVVEIQDRYDAAKEIE 226
Query: 473 KSLLELHQVFLDMAVMVEAQGEKMDDIEHHVLHSSHYVKDGTKNLQTAKMYQKSSRKWMC 294
KSLLELHQVFLDMAVMVE+QGE+MD+IEHHV+++SHYV DG L+TAK +Q++SRKWMC
Sbjct: 227 KSLLELHQVFLDMAVMVESQGEQMDEIEHHVINASHYVADGANELKTAKSHQRNSRKWMC 286
Query: 293 IGIIILLILILVIVIPIVTSLTSS 222
IGII+LL++IL++VIPI+TS +SS
Sbjct: 287 IGIIVLLLIILIVVIPIITSFSSS 310
>ref|NP_187788.1| syntaxin SYP121; protein id: At3g11820.1, supported by cDNA:
38899., supported by cDNA: gi_20260503, supported by
cDNA: gi_4206788 [Arabidopsis thaliana]
gi|28380149|sp|Q9ZSD4|S121_ARATH Syntaxin 121 (AtSYP121)
(Syntaxin-related protein At-Syr1)
gi|4206789|gb|AAD11809.1| syntaxin-related protein
At-SYR1 [Arabidopsis thaliana]
gi|6671938|gb|AAF23198.1|AC016795_11 putative syntaxin
[Arabidopsis thaliana] gi|20260504|gb|AAM13150.1|
putative syntaxin [Arabidopsis thaliana]
gi|21593428|gb|AAM65395.1| putative syntaxin
[Arabidopsis thaliana] gi|28059412|gb|AAO30056.1|
putative syntaxin [Arabidopsis thaliana]
Length = 346
Score = 169 bits (429), Expect = 2e-41
Identities = 79/144 (54%), Positives = 112/144 (76%)
Frame = -1
Query: 653 RYFTVTGENPDEEVIEKIISNGDEGGFMFLGKPVEEHGRGKVLETVAEIQDRHEGAKEVE 474
RYFTVTGENPDE ++++IS G+ FL K ++E GRG+VL+T+ EIQ+RH+ K++E
Sbjct: 171 RYFTVTGENPDERTLDRLISTGESE--RFLQKAIQEQGRGRVLDTINEIQERHDAVKDIE 228
Query: 473 KSLLELHQVFLDMAVMVEAQGEKMDDIEHHVLHSSHYVKDGTKNLQTAKMYQKSSRKWMC 294
K+L ELHQVFLDMAV+VE QG ++DDIE HV +S +++ GT LQTA++YQK++RKW C
Sbjct: 229 KNLRELHQVFLDMAVLVEHQGAQLDDIESHVGRASSFIRGGTDQLQTARVYQKNTRKWTC 288
Query: 293 IGIIILLILILVIVIPIVTSLTSS 222
I IIIL+I+I V+V+ ++ +S
Sbjct: 289 IAIIILIIIITVVVLAVLKPWNNS 312
>ref|NP_172591.1| syntaxin SYP125; protein id: At1g11250.1 [Arabidopsis thaliana]
gi|28380142|sp|Q9SXB0|S125_ARATH Putative syntaxin 125
(AtSYP125) gi|25402655|pir||D86246 hypothetical protein
[imported] - Arabidopsis thaliana
gi|5734739|gb|AAD50004.1|AC007259_17 Similar to syntaxin
[Arabidopsis thaliana]
Length = 298
Score = 161 bits (407), Expect = 8e-39
Identities = 71/138 (51%), Positives = 109/138 (78%)
Frame = -1
Query: 653 RYFTVTGENPDEEVIEKIISNGDEGGFMFLGKPVEEHGRGKVLETVAEIQDRHEGAKEVE 474
RYFT+TGE DE+ I+ +I++G+ F L K ++E GRG++L+T++EIQ+RH+ KE+E
Sbjct: 155 RYFTITGEKADEQTIDNLIASGESENF--LQKAIQEQGRGQILDTISEIQERHDAVKEIE 212
Query: 473 KSLLELHQVFLDMAVMVEAQGEKMDDIEHHVLHSSHYVKDGTKNLQTAKMYQKSSRKWMC 294
K+LLELHQVFLDMA +VEAQG+++++IE HV +S +V+ GT LQ A+ YQKSSRKW C
Sbjct: 213 KNLLELHQVFLDMAALVEAQGQQLNNIESHVAKASSFVRRGTDQLQDAREYQKSSRKWTC 272
Query: 293 IGIIILLILILVIVIPIV 240
II+ +++ ++++IP++
Sbjct: 273 YAIILFIVIFILLLIPLL 290
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 628,972,112
Number of Sequences: 1393205
Number of extensions: 17105138
Number of successful extensions: 348970
Number of sequences better than 10.0: 6517
Number of HSP's better than 10.0 without gapping: 104374
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 230834
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 28144814643
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)