Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
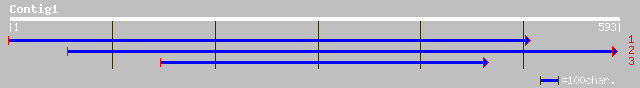
Query= KMC015904A_C01 KMC015904A_c01
(590 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sp|O04866|ARGD_ALNGL Acetylornithine aminotransferase, mitochond... 109 2e-23
ref|NP_178175.1| putative acetylornithine transaminase; protein ... 108 7e-23
gb|AAM63124.1| putative acetylornithine transaminase [Arabidopsi... 106 3e-22
ref|NP_764764.1| ornithine aminotransferase [Staphylococcus epid... 64 2e-09
ref|NP_276454.1| N-acetylornithine aminotransferase [Methanother... 64 2e-09
>sp|O04866|ARGD_ALNGL Acetylornithine aminotransferase, mitochondrial precursor (ACOAT)
(Acetylornithine transaminase) (AOTA)
gi|1944511|emb|CAA69936.1| acetylornithine
aminotransferase [Alnus glutinosa]
Length = 451
Score = 109 bits (273), Expect = 2e-23
Identities = 53/71 (74%), Positives = 64/71 (89%)
Frame = -2
Query: 586 HVKEIRGIGLIMGIDLDVPASPLVDACRDSGLLVLTAGKGNVVRLVPPLIITEQELEQQA 407
HV+E+RG+GLI+GI+LDV ASPLV+AC +SGLLVLTAGKGNVVR+VPPLIITEQELE+ A
Sbjct: 378 HVREVRGVGLIVGIELDVSASPLVNACLNSGLLVLTAGKGNVVRIVPPLIITEQELEKAA 437
Query: 406 DILCQAISVLD 374
+IL Q + LD
Sbjct: 438 EILLQCLPALD 448
>ref|NP_178175.1| putative acetylornithine transaminase; protein id: At1g80600.1,
supported by cDNA: 19586., supported by cDNA:
gi_15451027 [Arabidopsis thaliana]
gi|25406676|pir||B96838 hypothetical protein T21F11.7
[imported] - Arabidopsis thaliana
gi|6730727|gb|AAF27117.1|AC018849_5 putative
acetylornithine transaminase; 18117-19955 [Arabidopsis
thaliana] gi|15451028|gb|AAK96785.1| putative
acetylornithine transaminase [Arabidopsis thaliana]
gi|27311957|gb|AAO00944.1| putative acetylornithine
transaminase [Arabidopsis thaliana]
Length = 457
Score = 108 bits (269), Expect = 7e-23
Identities = 49/71 (69%), Positives = 64/71 (90%)
Frame = -2
Query: 586 HVKEIRGIGLIMGIDLDVPASPLVDACRDSGLLVLTAGKGNVVRLVPPLIITEQELEQQA 407
HVKE+RG GLI+G++LDVPAS LVDACRDSGLL+LTAGKGNVVR+VPPL+I+E+E+E+
Sbjct: 387 HVKEVRGEGLIIGVELDVPASSLVDACRDSGLLILTAGKGNVVRIVPPLVISEEEIERAV 446
Query: 406 DILCQAISVLD 374
+I+ Q ++ LD
Sbjct: 447 EIMSQNLTALD 457
>gb|AAM63124.1| putative acetylornithine transaminase [Arabidopsis thaliana]
Length = 457
Score = 106 bits (264), Expect = 3e-22
Identities = 48/71 (67%), Positives = 64/71 (89%)
Frame = -2
Query: 586 HVKEIRGIGLIMGIDLDVPASPLVDACRDSGLLVLTAGKGNVVRLVPPLIITEQELEQQA 407
+VKE+RG GLI+G++LDVPAS LVDACRDSGLL+LTAGKGNVVR+VPPL+I+E+E+E+
Sbjct: 387 YVKEVRGEGLIIGVELDVPASSLVDACRDSGLLILTAGKGNVVRIVPPLVISEEEIERAV 446
Query: 406 DILCQAISVLD 374
+I+ Q ++ LD
Sbjct: 447 EIMAQNLTALD 457
>ref|NP_764764.1| ornithine aminotransferase [Staphylococcus epidermidis ATCC 12228]
gi|27315673|gb|AAO04808.1|AE016748_42 ornithine
aminotransferase [Staphylococcus epidermidis ATCC 12228]
Length = 376
Score = 63.5 bits (153), Expect = 2e-09
Identities = 32/68 (47%), Positives = 51/68 (74%)
Frame = -2
Query: 589 RHVKEIRGIGLIMGIDLDVPASPLVDACRDSGLLVLTAGKGNVVRLVPPLIITEQELEQQ 410
R+V E+RG+GL++GI++ S +V + GL++LTAGK NV+RL+PPL IT+++LE+
Sbjct: 310 RNVIEVRGVGLMVGIEVTNDPSQVVREAKRMGLIILTAGK-NVIRLLPPLTITKKQLEKG 368
Query: 409 ADILCQAI 386
+IL + I
Sbjct: 369 IEILTEII 376
>ref|NP_276454.1| N-acetylornithine aminotransferase [Methanothermobacter
thermautotrophicus] gi|3913079|sp|O27392|ARGD_METTH
Acetylornithine aminotransferase (ACOAT)
gi|7434146|pir||G69044 N-acetylornithine
aminotransferase - Methanobacterium thermoautotrophicum
(strain Delta H) gi|2622444|gb|AAB85815.1|
N-acetylornithine aminotransferase [Methanothermobacter
thermautotrophicus str. Delta H]
Length = 390
Score = 63.5 bits (153), Expect = 2e-09
Identities = 32/70 (45%), Positives = 52/70 (73%), Gaps = 1/70 (1%)
Frame = -2
Query: 583 VKEIRGIGLIMGIDLDVPASPLVDACRDSGLLV-LTAGKGNVVRLVPPLIITEQELEQQA 407
V++IRG+GL++GI++D + +VDA R+ G+L+ TAGK V+R+VPPL+I ++E++
Sbjct: 323 VRDIRGVGLMIGIEIDGECAGVVDAAREMGVLINCTAGK--VIRIVPPLVIKKEEIDAAV 380
Query: 406 DILCQAISVL 377
D+L IS L
Sbjct: 381 DVLGHVISDL 390
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 486,273,411
Number of Sequences: 1393205
Number of extensions: 10313357
Number of successful extensions: 24794
Number of sequences better than 10.0: 360
Number of HSP's better than 10.0 without gapping: 23944
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24659
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 22569056698
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)