Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC015891A_C01 KMC015891A_c01
(606 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
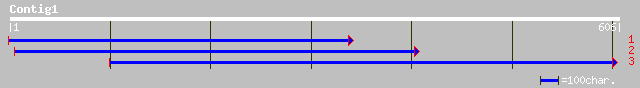
gb|AAM61682.1| unknown [Arabidopsis thaliana] 241 7e-63
gb|AAL75891.1| At1g21480/F24J8_23 [Arabidopsis thaliana] 241 7e-63
ref|NP_564141.1| Exostosin family; protein id: At1g21480.1, supp... 241 7e-63
pir||G86347 hypothetical protein F24J8.10 - Arabidopsis thaliana... 187 7e-47
ref|NP_564443.1| Exostosin family; protein id: At1g34270.1, supp... 71 1e-11
>gb|AAM61682.1| unknown [Arabidopsis thaliana]
Length = 462
Score = 241 bits (614), Expect = 7e-63
Identities = 109/147 (74%), Positives = 125/147 (84%)
Frame = -3
Query: 604 LRNSKFCLAPRGESSWTLRFYESFFVECVPVIISDQIELPFQNVIDYSQISIKWPSNRIG 425
LRN+KFCLAPRGESSWTLRFYESFFVECVPV++SD ELPFQNVIDY+Q+SIKWPS RIG
Sbjct: 316 LRNAKFCLAPRGESSWTLRFYESFFVECVPVLLSDHAELPFQNVIDYAQVSIKWPSTRIG 375
Query: 424 PELLQYLESIPDQNIEAIIARGRKVRCFWIYASDLEPCSAMRGIMWELQRKVRQFHQSTE 245
E L YL SI D++IE +IARGRK+RC ++Y D PCSA++GI+WELQRKVR F QSTE
Sbjct: 376 SEFLDYLASISDRDIEGMIARGRKIRCLFVYGPDSAPCSAVKGILWELQRKVRHFQQSTE 435
Query: 244 TFWLHNGSLVNRNLVEFSKWELPVSLP 164
TFWLHNGS VNR LV+FS W P+ LP
Sbjct: 436 TFWLHNGSFVNRELVQFSSWRPPMPLP 462
>gb|AAL75891.1| At1g21480/F24J8_23 [Arabidopsis thaliana]
Length = 462
Score = 241 bits (614), Expect = 7e-63
Identities = 109/147 (74%), Positives = 125/147 (84%)
Frame = -3
Query: 604 LRNSKFCLAPRGESSWTLRFYESFFVECVPVIISDQIELPFQNVIDYSQISIKWPSNRIG 425
LRN+KFCLAPRGESSWTLRFYESFFVECVPV++SD ELPFQNVIDY+Q+SIKWPS RIG
Sbjct: 316 LRNAKFCLAPRGESSWTLRFYESFFVECVPVLLSDHAELPFQNVIDYAQVSIKWPSTRIG 375
Query: 424 PELLQYLESIPDQNIEAIIARGRKVRCFWIYASDLEPCSAMRGIMWELQRKVRQFHQSTE 245
E L YL SI D++IE +IARGRK+RC ++Y D PCSA++GI+WELQRKVR F QSTE
Sbjct: 376 SEFLDYLASISDRDIEGMIARGRKIRCLFVYGPDSAPCSAVKGILWELQRKVRHFQQSTE 435
Query: 244 TFWLHNGSLVNRNLVEFSKWELPVSLP 164
TFWLHNGS VNR LV+FS W P+ LP
Sbjct: 436 TFWLHNGSFVNRELVQFSSWRPPMPLP 462
>ref|NP_564141.1| Exostosin family; protein id: At1g21480.1, supported by cDNA:
1324., supported by cDNA: gi_18650642 [Arabidopsis
thaliana]
Length = 462
Score = 241 bits (614), Expect = 7e-63
Identities = 109/147 (74%), Positives = 125/147 (84%)
Frame = -3
Query: 604 LRNSKFCLAPRGESSWTLRFYESFFVECVPVIISDQIELPFQNVIDYSQISIKWPSNRIG 425
LRN+KFCLAPRGESSWTLRFYESFFVECVPV++SD ELPFQNVIDY+Q+SIKWPS RIG
Sbjct: 316 LRNAKFCLAPRGESSWTLRFYESFFVECVPVLLSDHAELPFQNVIDYAQVSIKWPSTRIG 375
Query: 424 PELLQYLESIPDQNIEAIIARGRKVRCFWIYASDLEPCSAMRGIMWELQRKVRQFHQSTE 245
E L YL SI D++IE +IARGRK+RC ++Y D PCSA++GI+WELQRKVR F QSTE
Sbjct: 376 SEFLDYLASISDRDIEGMIARGRKIRCLFVYGPDSAPCSAVKGILWELQRKVRHFQQSTE 435
Query: 244 TFWLHNGSLVNRNLVEFSKWELPVSLP 164
TFWLHNGS VNR LV+FS W P+ LP
Sbjct: 436 TFWLHNGSFVNRELVQFSSWRPPMPLP 462
>pir||G86347 hypothetical protein F24J8.10 - Arabidopsis thaliana
gi|9454585|gb|AAF87908.1|AC015447_18 Hypothetical
protein [Arabidopsis thaliana]
Length = 414
Score = 187 bits (476), Expect = 7e-47
Identities = 84/121 (69%), Positives = 99/121 (81%)
Frame = -3
Query: 526 ECVPVIISDQIELPFQNVIDYSQISIKWPSNRIGPELLQYLESIPDQNIEAIIARGRKVR 347
ECVPV++SD ELPFQNVIDY+Q+SIKWPS RIG E L YL SI D++IE +IARGRK+R
Sbjct: 294 ECVPVLLSDHAELPFQNVIDYAQVSIKWPSTRIGSEFLDYLASISDRDIEGMIARGRKIR 353
Query: 346 CFWIYASDLEPCSAMRGIMWELQRKVRQFHQSTETFWLHNGSLVNRNLVEFSKWELPVSL 167
C ++Y D PCSA++GI+WELQRKVR F QSTETFWLHNGS VNR LV+FS W P+ L
Sbjct: 354 CLFVYGPDSAPCSAVKGILWELQRKVRHFQQSTETFWLHNGSFVNRELVQFSSWRPPMPL 413
Query: 166 P 164
P
Sbjct: 414 P 414
>ref|NP_564443.1| Exostosin family; protein id: At1g34270.1, supported by cDNA:
gi_15450927, supported by cDNA: gi_20148710 [Arabidopsis
thaliana] gi|25403422|pir||H86466 protein F23M19.7
[imported] - Arabidopsis thaliana
gi|5091619|gb|AAD39607.1|AC007454_6 F23M19.7
[Arabidopsis thaliana] gi|15450928|gb|AAK96735.1|
Unknown protein [Arabidopsis thaliana]
gi|20148711|gb|AAM10246.1| unknown protein [Arabidopsis
thaliana]
Length = 477
Score = 70.9 bits (172), Expect = 1e-11
Identities = 41/121 (33%), Positives = 71/121 (57%), Gaps = 9/121 (7%)
Frame = -3
Query: 604 LRNSKFCLAPRGESSWTLRFYESFFVECVPVIISDQIELPFQNVIDYSQISI-KWPSNRI 428
+RNS+FCL P G++ + R +++ C+PVI+SD IELPF+ +IDYS+ S+ S+ +
Sbjct: 331 MRNSEFCLHPAGDTPTSCRLFDAIQSLCIPVIVSDTIELPFEGIIDYSEFSVFASVSDAL 390
Query: 427 GPE-LLQYLESIPDQNIEAIIARGRKVRCFWIY-------ASDLEPCSAMRGIMWELQRK 272
P+ L +L ++ E + +R KV+ ++Y +EP A+ I ++Q+K
Sbjct: 391 TPKWLANHLGRFSEREKETLRSRIAKVQSVFVYDNGHADGIGPIEPNGAVNHIWKKVQQK 450
Query: 271 V 269
V
Sbjct: 451 V 451
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 521,428,136
Number of Sequences: 1393205
Number of extensions: 10962867
Number of successful extensions: 26746
Number of sequences better than 10.0: 99
Number of HSP's better than 10.0 without gapping: 25782
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26722
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23997478008
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)