Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC015862A_C01 KMC015862A_c01
(748 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
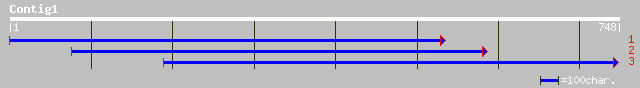
ref|NP_190346.2| putative protein; protein id: At3g47620.1, supp... 50 2e-07
pir||T45722 hypothetical protein F1P2.170 - Arabidopsis thaliana... 50 2e-07
ref|NP_564973.1| expressed protein; protein id: At1g69690.1, sup... 47 4e-04
ref|NP_700532.1| hypothetical protein [Plasmodium falciparum 3D7... 39 0.060
ref|NP_701177.1| structural maintenance of chromosome protein, p... 38 0.17
>ref|NP_190346.2| putative protein; protein id: At3g47620.1, supported by cDNA:
gi_16604510 [Arabidopsis thaliana]
gi|16604511|gb|AAL24261.1| AT3g47620/F1P2_170
[Arabidopsis thaliana] gi|21655289|gb|AAM65356.1|
AT3g47620/F1P2_170 [Arabidopsis thaliana]
Length = 489
Score = 49.7 bits (117), Expect(2) = 2e-07
Identities = 40/109 (36%), Positives = 48/109 (43%), Gaps = 37/109 (33%)
Frame = -1
Query: 748 GSYFLQSSSVGTIPASHA--QIPASIWMLA--------NSNNNQ-----------GMSGG 632
G Y LQSS+ G+ + A QIP + WM+A NNNQ G GG
Sbjct: 305 GGYTLQSSNSGSTATAAAAQQIPGNFWMVAAAAAAGGGGGNNNQTGGLMTASIGTGGGGG 364
Query: 631 DWV----------------NHSGLYRGALQSGLHFMNFPTHMALLPGQQ 533
+ V SG+ GA+ SGLHFMNF MA L GQQ
Sbjct: 365 EPVWTFPSINTAAAALYRSGVSGVPSGAVSSGLHFMNFAAPMAFLTGQQ 413
Score = 27.3 bits (59), Expect(2) = 2e-07
Identities = 11/38 (28%), Positives = 20/38 (51%), Gaps = 3/38 (7%)
Frame = -2
Query: 441 VSESQASGSQSHHDGGGEDDPRENNNHH---HHHS*MS 337
++E + DGGG+ + ++HH HHH+ +S
Sbjct: 423 INEDSNNNEGGRSDGGGDHHNTQRHHHHQQQHHHNILS 460
>pir||T45722 hypothetical protein F1P2.170 - Arabidopsis thaliana
gi|6522545|emb|CAB61988.1| putative protein [Arabidopsis
thaliana]
Length = 477
Score = 49.7 bits (117), Expect(2) = 2e-07
Identities = 40/109 (36%), Positives = 48/109 (43%), Gaps = 37/109 (33%)
Frame = -1
Query: 748 GSYFLQSSSVGTIPASHA--QIPASIWMLA--------NSNNNQ-----------GMSGG 632
G Y LQSS+ G+ + A QIP + WM+A NNNQ G GG
Sbjct: 293 GGYTLQSSNSGSTATAAAAQQIPGNFWMVAAAAAAGGGGGNNNQTGGLMTASIGTGGGGG 352
Query: 631 DWV----------------NHSGLYRGALQSGLHFMNFPTHMALLPGQQ 533
+ V SG+ GA+ SGLHFMNF MA L GQQ
Sbjct: 353 EPVWTFPSINTAAAALYRSGVSGVPSGAVSSGLHFMNFAAPMAFLTGQQ 401
Score = 27.3 bits (59), Expect(2) = 2e-07
Identities = 11/38 (28%), Positives = 20/38 (51%), Gaps = 3/38 (7%)
Frame = -2
Query: 441 VSESQASGSQSHHDGGGEDDPRENNNHH---HHHS*MS 337
++E + DGGG+ + ++HH HHH+ +S
Sbjct: 411 INEDSNNNEGGRSDGGGDHHNTQRHHHHQQQHHHNILS 448
>ref|NP_564973.1| expressed protein; protein id: At1g69690.1, supported by cDNA:
gi_15912212, supported by cDNA: gi_19547990 [Arabidopsis
thaliana] gi|25404829|pir||G96718 unknown protein,
54453-53476 [imported] - Arabidopsis thaliana
gi|12325189|gb|AAG52540.1|AC013289_7 unknown protein;
54453-53476 [Arabidopsis thaliana]
gi|15912213|gb|AAL08240.1| At1g69690/T6C23_11
[Arabidopsis thaliana] gi|19547991|gb|AAL87359.1|
At1g69690/T6C23_11 [Arabidopsis thaliana]
Length = 325
Score = 46.6 bits (109), Expect = 4e-04
Identities = 32/90 (35%), Positives = 45/90 (49%), Gaps = 11/90 (12%)
Frame = -1
Query: 748 GSYFLQSSSVGTIPASHAQIPASIWMLANSNNN----------QGMSGGDWVN-HSGLYR 602
G+Y +QS++ G++P S + A W ++ N G+ GD N +SG
Sbjct: 184 GNYLVQSTA-GSLPTSQSPATAPFWSSGDNTQNLWAFNINPHHSGVVAGDVYNPNSGGSG 242
Query: 601 GALQSGLHFMNFPTHMALLPGQQLGSGIGG 512
G SG+H MNF +AL GQ L SG GG
Sbjct: 243 GG--SGVHLMNFAAPIALFSGQPLASGYGG 270
>ref|NP_700532.1| hypothetical protein [Plasmodium falciparum 3D7]
gi|23494921|gb|AAN35256.1|AE014829_56 hypothetical
protein [Plasmodium falciparum 3D7]
Length = 1564
Score = 39.3 bits (90), Expect = 0.060
Identities = 31/110 (28%), Positives = 48/110 (43%)
Frame = +3
Query: 51 EINQRKLFFSLQKKTPAKNPTVKQYHHLSETLKNLPHLFFHHARTSSKTLINHTKKEINY 230
EI +RKL F KK +QY S + H + N+ K+INY
Sbjct: 1117 EILKRKLLFEENKKKEYYEEMEEQYISSSSSSSLNVHKY------------NNECKDINY 1164
Query: 231 SFKINIKQNSISLQGNQTSHVELTNPSNYYKINKKNSSMSDDDDDCCSHE 380
F+ N +NS+ +V++T P + IN+K +DDD+ H+
Sbjct: 1165 PFETN--KNSLPNYTKDKQNVDITEPKTQH-INQKKKHFTDDDNISSVHD 1211
>ref|NP_701177.1| structural maintenance of chromosome protein, putative [Plasmodium
falciparum 3D7] gi|23496242|gb|AAN35901.1|AE014840_49
structural maintenance of chromosome protein, putative
[Plasmodium falciparum 3D7]
Length = 1818
Score = 37.7 bits (86), Expect = 0.17
Identities = 37/137 (27%), Positives = 54/137 (39%), Gaps = 11/137 (8%)
Frame = +3
Query: 6 KVNYCTDNDDKISCNEINQRKLFF--SLQKKTPAKNPTVKQYHHLSETLKNLPHLFFHHA 179
K NY N KI NEI+++K F SLQK K + H ++ ++NL
Sbjct: 576 KKNYVQINQLKILLNEISEKKKFCNDSLQKLVSNKKLKINMQEHCNKFMENLNLQIKEQN 635
Query: 180 RTSSKTLINHTKKEINYSFKINIKQNSISL---QGNQTSHVEL--TNPSNYY----KINK 332
+ N K I + K QN I + SH++ N SN + I K
Sbjct: 636 KKLENEYKNKLKINIKFFSKYECVQNIIKTKIEECKMNSHIDQQDQNESNVHLNNENIKK 695
Query: 333 KNSSMSDDDDDCCSHED 383
KN + +H+D
Sbjct: 696 KNDKKGKKKLNINNHDD 712
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 668,053,376
Number of Sequences: 1393205
Number of extensions: 15822252
Number of successful extensions: 69735
Number of sequences better than 10.0: 81
Number of HSP's better than 10.0 without gapping: 55657
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 65493
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 36032594816
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)