Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC015821A_C01 KMC015821A_c01
(658 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
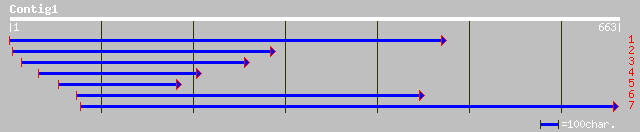
ref|NP_565099.1| expressed protein; protein id: At1g75080.1, sup... 121 9e-27
gb|AAL57684.1| At1g75080/F9E10_7 [Arabidopsis thaliana] gi|20147... 121 9e-27
pir||G96780 unknown protein F9E10.7 [imported] - Arabidopsis tha... 121 9e-27
ref|NP_564081.1| expressed protein; protein id: At1g19350.1, sup... 115 5e-25
gb|AAM64408.1| unknown [Arabidopsis thaliana] 115 5e-25
>ref|NP_565099.1| expressed protein; protein id: At1g75080.1, supported by cDNA:
33367., supported by cDNA: gi_18086460, supported by
cDNA: gi_20147314 [Arabidopsis thaliana]
gi|20270971|gb|AAM18490.1|AF494338_1 BZR1 protein
[Arabidopsis thaliana] gi|21592862|gb|AAM64812.1|
unknown [Arabidopsis thaliana]
Length = 336
Score = 121 bits (303), Expect = 9e-27
Identities = 62/110 (56%), Positives = 77/110 (69%), Gaps = 2/110 (1%)
Frame = -2
Query: 657 PSHRH-LYTPPTIPECDESDSSTVESGQWLNFQAFAPFAPSPPSGMQMSPTFNFIKPVMR 481
P+HRH +TP TIPECDESDSSTV+SG W++FQ FA P S + SPTFN +KP +
Sbjct: 225 PTHRHQFHTPATIPECDESDSSTVDSGHWISFQKFAQQQPFSASMVPTSPTFNLVKPAPQ 284
Query: 480 QQQNMPDSRDQEMRSSEADF-GVQVKPWVGERIHEVGLDDLELTLGSGKA 334
Q + + +SSE F QVKPW GERIH+VG++DLELTLG+GKA
Sbjct: 285 QMSPNTAAFQEIGQSSEFKFENSQVKPWEGERIHDVGMEDLELTLGNGKA 334
>gb|AAL57684.1| At1g75080/F9E10_7 [Arabidopsis thaliana] gi|20147315|gb|AAM10371.1|
At1g75080/F9E10_7 [Arabidopsis thaliana]
Length = 336
Score = 121 bits (303), Expect = 9e-27
Identities = 62/110 (56%), Positives = 77/110 (69%), Gaps = 2/110 (1%)
Frame = -2
Query: 657 PSHRH-LYTPPTIPECDESDSSTVESGQWLNFQAFAPFAPSPPSGMQMSPTFNFIKPVMR 481
P+HRH +TP TIPECDESDSSTV+SG W++FQ FA P S + SPTFN +KP +
Sbjct: 225 PTHRHQFHTPATIPECDESDSSTVDSGHWISFQKFAQQQPFSASMVPTSPTFNLVKPAPQ 284
Query: 480 QQQNMPDSRDQEMRSSEADF-GVQVKPWVGERIHEVGLDDLELTLGSGKA 334
Q + + +SSE F QVKPW GERIH+VG++DLELTLG+GKA
Sbjct: 285 QMSPNTAAFQEIGQSSEFKFENSQVKPWEGERIHDVGMEDLELTLGNGKA 334
>pir||G96780 unknown protein F9E10.7 [imported] - Arabidopsis thaliana
gi|12323903|gb|AAG51929.1|AC013258_23 unknown protein;
17658-16304 [Arabidopsis thaliana]
Length = 333
Score = 121 bits (303), Expect = 9e-27
Identities = 62/110 (56%), Positives = 77/110 (69%), Gaps = 2/110 (1%)
Frame = -2
Query: 657 PSHRH-LYTPPTIPECDESDSSTVESGQWLNFQAFAPFAPSPPSGMQMSPTFNFIKPVMR 481
P+HRH +TP TIPECDESDSSTV+SG W++FQ FA P S + SPTFN +KP +
Sbjct: 222 PTHRHQFHTPATIPECDESDSSTVDSGHWISFQKFAQQQPFSASMVPTSPTFNLVKPAPQ 281
Query: 480 QQQNMPDSRDQEMRSSEADF-GVQVKPWVGERIHEVGLDDLELTLGSGKA 334
Q + + +SSE F QVKPW GERIH+VG++DLELTLG+GKA
Sbjct: 282 QMSPNTAAFQEIGQSSEFKFENSQVKPWEGERIHDVGMEDLELTLGNGKA 331
>ref|NP_564081.1| expressed protein; protein id: At1g19350.1, supported by cDNA:
24157., supported by cDNA: gi_18086445, supported by
cDNA: gi_6651068 [Arabidopsis thaliana]
gi|25352560|pir||G86326 protein F18O14.7 [imported] -
Arabidopsis thaliana gi|8778414|gb|AAF79422.1|AC025808_4
F18O14.7 [Arabidopsis thaliana]
gi|18086446|gb|AAL57677.1| At1g19350/F18O14_4
[Arabidopsis thaliana] gi|20908096|tpg|DAA00023.1| TPA:
BZR2; BES1 [Arabidopsis thaliana]
Length = 335
Score = 115 bits (288), Expect = 5e-25
Identities = 59/109 (54%), Positives = 73/109 (66%), Gaps = 1/109 (0%)
Frame = -2
Query: 651 HRHLYTPPTIPECDESDSSTVESGQWLNFQAFAPFAPSPPSGMQMSPTFNFIKPVMRQQQ 472
HR + P TIPECDESDSSTV+SG W++FQ FA P S + SPTFN +KP +Q
Sbjct: 227 HRQFHAPATIPECDESDSSTVDSGHWISFQKFAQQQPFSASMVPTSPTFNLVKPAPQQLS 286
Query: 471 NMPDSRDQEMRSSEADF-GVQVKPWVGERIHEVGLDDLELTLGSGKAPS 328
+ + +SSE F QVKPW GERIH+V ++DLELTLG+GKA S
Sbjct: 287 PNTAAIQEIGQSSEFKFENSQVKPWEGERIHDVAMEDLELTLGNGKAHS 335
>gb|AAM64408.1| unknown [Arabidopsis thaliana]
Length = 335
Score = 115 bits (288), Expect = 5e-25
Identities = 59/109 (54%), Positives = 73/109 (66%), Gaps = 1/109 (0%)
Frame = -2
Query: 651 HRHLYTPPTIPECDESDSSTVESGQWLNFQAFAPFAPSPPSGMQMSPTFNFIKPVMRQQQ 472
HR + P TIPECDESDSSTV+SG W++FQ FA P S + SPTFN +KP +Q
Sbjct: 227 HRQFHAPATIPECDESDSSTVDSGHWISFQKFAQQQPFSASMVPTSPTFNLVKPAPQQLS 286
Query: 471 NMPDSRDQEMRSSEADF-GVQVKPWVGERIHEVGLDDLELTLGSGKAPS 328
+ + +SSE F QVKPW GERIH+V ++DLELTLG+GKA S
Sbjct: 287 PNTAAIQEIGQSSEFKFENSQVKPWEGERIHDVAMEDLELTLGNGKAHS 335
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 584,708,758
Number of Sequences: 1393205
Number of extensions: 13328303
Number of successful extensions: 55451
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 42178
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 53066
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 28006887348
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)