Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC015709A_C01 KMC015709A_c01
(629 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
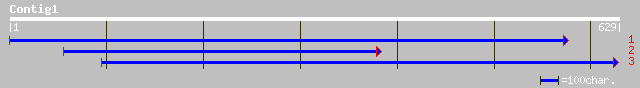
sp|Q43111|PME3_PHAVU Pectinesterase 3 precursor (Pectin methyles... 246 2e-64
gb|AAM65650.1| pectinesterase, putative [Arabidopsis thaliana] 189 3e-47
ref|NP_175787.1| pectinesterase family; protein id: At1g53840.1,... 189 3e-47
ref|NP_196115.1| pectinesterase family; protein id: At5g04960.1 ... 169 4e-41
ref|NP_188047.1| putative pectin methylesterase; protein id: At3... 168 6e-41
>sp|Q43111|PME3_PHAVU Pectinesterase 3 precursor (Pectin methylesterase 3) (PE 3)
gi|1076515|pir||S53105 pectinesterase precursor - kidney
bean gi|732913|emb|CAA59482.1| pectinesterase [Phaseolus
vulgaris]
Length = 581
Score = 246 bits (628), Expect = 2e-64
Identities = 112/146 (76%), Positives = 128/146 (86%)
Frame = -3
Query: 627 CKIMPRQPMPNQFYTITAQGKKDPNQNTGIVIQKSTITPLESTYTAPTYLGRPWKDFSTT 448
CKIMPRQP+PNQF TITAQGKKDPNQNTGI+IQKSTITP + TAPTYLGRPWKDFSTT
Sbjct: 435 CKIMPRQPLPNQFNTITAQGKKDPNQNTGIIIQKSTITPFGNNLTAPTYLGRPWKDFSTT 494
Query: 447 LIMQSEIGSFLKPVGWVSWVANVDPPATIFYAEHQNSGPGSDVAQRVKWTGYKSTLTADD 268
+IMQS+IG+ L PVGW+SWV NV+PP TIFYAE+QNSGPG+DV+QRVKW GYK T+T +
Sbjct: 495 VIMQSDIGALLNPVGWMSWVPNVEPPTTIFYAEYQNSGPGADVSQRVKWAGYKPTITDRN 554
Query: 267 LGKFTLASFIQGPEWLPDTAVAFDST 190
+FT+ SFIQGPEWLP+ AV FDST
Sbjct: 555 AEEFTVQSFIQGPEWLPNAAVQFDST 580
>gb|AAM65650.1| pectinesterase, putative [Arabidopsis thaliana]
Length = 586
Score = 189 bits (480), Expect = 3e-47
Identities = 84/141 (59%), Positives = 111/141 (78%)
Frame = -3
Query: 627 CKIMPRQPMPNQFYTITAQGKKDPNQNTGIVIQKSTITPLESTYTAPTYLGRPWKDFSTT 448
CKIMPRQP+ NQF TITAQGKKDPNQ++G+ IQ+ TI+ APTYLGRPWK+FSTT
Sbjct: 442 CKIMPRQPLSNQFNTITAQGKKDPNQSSGMSIQRCTIST-NGNVIAPTYLGRPWKEFSTT 500
Query: 447 LIMQSEIGSFLKPVGWVSWVANVDPPATIFYAEHQNSGPGSDVAQRVKWTGYKSTLTADD 268
+IM++ IG+ ++P GW+SWV+ VDPPA+I Y E++N+GPGSDV QRVKW GYK ++ +
Sbjct: 501 VIMETVIGAVVRPSGWMSWVSGVDPPASIVYGEYKNTGPGSDVTQRVKWAGYKPVMSDAE 560
Query: 267 LGKFTLASFIQGPEWLPDTAV 205
KFT+A+ + G +W+P T V
Sbjct: 561 AAKFTVATLLHGADWIPATGV 581
>ref|NP_175787.1| pectinesterase family; protein id: At1g53840.1, supported by cDNA:
41374., supported by cDNA: gi_15809859 [Arabidopsis
thaliana] gi|6093736|sp|Q43867|PME1_ARATH Pectinesterase
1 precursor (Pectin methylesterase 1) (PE 1)
gi|2129666|pir||JC4778 pectinesterase (EC 3.1.1.11) PME1
precursor - Arabidopsis thaliana
gi|550306|emb|CAA57275.1| ATPME1 [Arabidopsis thaliana]
gi|903895|gb|AAC50024.1| ATPME1 precursor [Arabidopsis
thaliana] gi|6056393|gb|AAF02857.1|AC009324_6
Pectinesterase 1 [Arabidopsis thaliana]
gi|15809860|gb|AAL06858.1| At1g53840/T18A20_7
[Arabidopsis thaliana]
Length = 586
Score = 189 bits (480), Expect = 3e-47
Identities = 84/141 (59%), Positives = 111/141 (78%)
Frame = -3
Query: 627 CKIMPRQPMPNQFYTITAQGKKDPNQNTGIVIQKSTITPLESTYTAPTYLGRPWKDFSTT 448
CKIMPRQP+ NQF TITAQGKKDPNQ++G+ IQ+ TI+ APTYLGRPWK+FSTT
Sbjct: 442 CKIMPRQPLSNQFNTITAQGKKDPNQSSGMSIQRCTISA-NGNVIAPTYLGRPWKEFSTT 500
Query: 447 LIMQSEIGSFLKPVGWVSWVANVDPPATIFYAEHQNSGPGSDVAQRVKWTGYKSTLTADD 268
+IM++ IG+ ++P GW+SWV+ VDPPA+I Y E++N+GPGSDV QRVKW GYK ++ +
Sbjct: 501 VIMETVIGAVVRPSGWMSWVSGVDPPASIVYGEYKNTGPGSDVTQRVKWAGYKPVMSDAE 560
Query: 267 LGKFTLASFIQGPEWLPDTAV 205
KFT+A+ + G +W+P T V
Sbjct: 561 AAKFTVATLLHGADWIPATGV 581
>ref|NP_196115.1| pectinesterase family; protein id: At5g04960.1 [Arabidopsis
thaliana] gi|10178035|dbj|BAB11518.1| pectinesterase
[Arabidopsis thaliana]
Length = 564
Score = 169 bits (427), Expect = 4e-41
Identities = 80/145 (55%), Positives = 100/145 (68%)
Frame = -3
Query: 627 CKIMPRQPMPNQFYTITAQGKKDPNQNTGIVIQKSTITPLESTYTAPTYLGRPWKDFSTT 448
C+I+PR+PM Q TITAQG+KDPNQNTGI I TI PL++ T+LGRPWKDFSTT
Sbjct: 419 CEILPRRPMKGQQNTITAQGRKDPNQNTGISIHNCTIKPLDNLTDIQTFLGRPWKDFSTT 478
Query: 447 LIMQSEIGSFLKPVGWVSWVANVDPPATIFYAEHQNSGPGSDVAQRVKWTGYKSTLTADD 268
+IM+S + F+ P GW+ W + P TIFYAE+ NSGPG+ RVKW G K++LT +
Sbjct: 479 VIMKSFMDKFINPKGWLPWTGDT-APDTIFYAEYLNSGPGASTKNRVKWQGLKTSLTKKE 537
Query: 267 LGKFTLASFIQGPEWLPDTAVAFDS 193
KFT+ FI G WLP T V F+S
Sbjct: 538 ANKFTVKPFIDGNNWLPATKVPFNS 562
>ref|NP_188047.1| putative pectin methylesterase; protein id: At3g14300.1 [Arabidopsis
thaliana] gi|9279578|dbj|BAB01036.1| pectinesterase-like
protein [Arabidopsis thaliana]
Length = 968
Score = 168 bits (425), Expect = 6e-41
Identities = 79/139 (56%), Positives = 103/139 (73%), Gaps = 2/139 (1%)
Frame = -3
Query: 627 CKIMPRQPMPNQFYTITAQGKKDPNQNTGIVIQKSTITPLESTYTAPTYLGRPWKDFSTT 448
C I PRQP+PNQF TITA+G ++ NQNTGI I + TI+P TA TYLGRPWK FS T
Sbjct: 822 CSIRPRQPLPNQFNTITAEGTQEANQNTGISIHQCTISP-NGNVTATTYLGRPWKLFSKT 880
Query: 447 LIMQSEIGSFLKPVGWVSWVANVD-PPATIFYAEHQNSGPGSDVAQRVKWTGYKSTLTAD 271
+IMQS IGSF+ P GW++W + D PP TIFY E++NSGPGSD+++RVKW GYK + D
Sbjct: 881 VIMQSVIGSFVNPAGWIAWNSTYDPPPRTIFYREYKNSGPGSDLSKRVKWAGYKPISSDD 940
Query: 270 DLGKFTLASFIQGPE-WLP 217
+ +FT+ F++G + W+P
Sbjct: 941 EAARFTVKYFLRGDDNWIP 959
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 613,666,451
Number of Sequences: 1393205
Number of extensions: 14377310
Number of successful extensions: 49693
Number of sequences better than 10.0: 222
Number of HSP's better than 10.0 without gapping: 47888
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 49299
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25870486187
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)