Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC015704A_C01 KMC015704A_c01
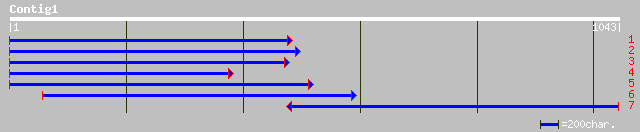
(1043 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAB64536.1| ferritin [Glycine max] 405 e-112
pir||T08124 ferritin 2 precursor - cowpea gi|2970654|gb|AAC06027... 365 e-100
gb|AAN06322.1| ferritin 2 [Nicotiana tabacum] 362 4e-99
gb|AAM11429.1| ferritin [Nicotiana tabacum] 362 7e-99
dbj|BAB64537.1| ferritin [Glycine max] 356 3e-97
>dbj|BAB64536.1| ferritin [Glycine max]
Length = 256
Score = 405 bits (1041), Expect = e-112
Identities = 212/257 (82%), Positives = 226/257 (87%), Gaps = 9/257 (3%)
Frame = -3
Query: 966 MLLRAASSFSLLNSNGGDHLLPPLFSSSVS--------PKTFLPCATKGSNNRPLTGVVF 811
MLLR ASSFSLL +N DH+LP SSS K +PCATK +NNRPLTGVVF
Sbjct: 1 MLLRTASSFSLLKANA-DHILPLPNSSSSGIIRYSQSLGKNLVPCATKDTNNRPLTGVVF 59
Query: 810 EPFEEVKKELDLVPTVPQASLARQKFVDECEAAINEQINVEYTVSYVYHAMFAYFDRDNV 631
EPFEEVKKELDLVPTVPQASLARQK+ D+CEA INEQINVEY VSYVYHAMFAYFDRDNV
Sbjct: 60 EPFEEVKKELDLVPTVPQASLARQKYTDDCEATINEQINVEYNVSYVYHAMFAYFDRDNV 119
Query: 630 ALKGLAKFFKESSMEEREHAEKLMEYQNKRGGRVKLQSIVMPPSEFEHEEKGDALYAMEL 451
ALKGLAKFFKESS EEREHAEKLMEYQNKRGG+VKLQSIVMP SEF+HEEKGDALYAMEL
Sbjct: 120 ALKGLAKFFKESSEEEREHAEKLMEYQNKRGGKVKLQSIVMPLSEFDHEEKGDALYAMEL 179
Query: 450 ALSLEKLTNEKLLNLHSVASKNNDAQLTDFIESEYLGEQVESIKPISEYVAQLRRVGQGH 271
ALSLEKLTNEKLLNLHSVASKNND QL DFIESE+LGEQVE+IK ISEYVAQLRRVG+GH
Sbjct: 180 ALSLEKLTNEKLLNLHSVASKNNDVQLADFIESEFLGEQVEAIKKISEYVAQLRRVGKGH 239
Query: 270 GTWHFNQMLL-QDGVAA 223
G WHF+QMLL ++GVAA
Sbjct: 240 GVWHFDQMLLHEEGVAA 256
>pir||T08124 ferritin 2 precursor - cowpea gi|2970654|gb|AAC06027.1| ferritin
subunit cowpea2 precursor [Vigna unguiculata]
Length = 250
Score = 365 bits (937), Expect = e-100
Identities = 191/248 (77%), Positives = 214/248 (86%), Gaps = 3/248 (1%)
Frame = -3
Query: 966 MLLR--AASSFSLLNSNGGDHLLPPLFSSSVSPKTFLPCATKGS-NNRPLTGVVFEPFEE 796
MLLR AASS SL N N P+ +++ S + A KGS N+R LTGV+FEPFEE
Sbjct: 1 MLLRTAAASSLSLFNPNAEPSRSVPVLANNAS--RLVVRAAKGSTNHRALTGVIFEPFEE 58
Query: 795 VKKELDLVPTVPQASLARQKFVDECEAAINEQINVEYTVSYVYHAMFAYFDRDNVALKGL 616
VKKELDLVPTVPQASLARQK+VDE EAA+NEQINVEY VSYVYHA+FAYFDRDNVAL+GL
Sbjct: 59 VKKELDLVPTVPQASLARQKYVDESEAAVNEQINVEYNVSYVYHALFAYFDRDNVALRGL 118
Query: 615 AKFFKESSMEEREHAEKLMEYQNKRGGRVKLQSIVMPPSEFEHEEKGDALYAMELALSLE 436
AKFFKESS EEREHAEKLMEYQN+RGG+VKLQSIVMP SEF+H +KGDAL+AMELALSLE
Sbjct: 119 AKFFKESSEEEREHAEKLMEYQNRRGGKVKLQSIVMPLSEFDHADKGDALHAMELALSLE 178
Query: 435 KLTNEKLLNLHSVASKNNDAQLTDFIESEYLGEQVESIKPISEYVAQLRRVGQGHGTWHF 256
KLTNEKLL+LHSVA+KN D QL DF+ESE+LGEQVESIK ISEYVAQLRRVG+GHG WHF
Sbjct: 179 KLTNEKLLHLHSVATKNGDVQLADFVESEFLGEQVESIKRISEYVAQLRRVGKGHGVWHF 238
Query: 255 NQMLLQDG 232
+QMLL +G
Sbjct: 239 DQMLLHEG 246
>gb|AAN06322.1| ferritin 2 [Nicotiana tabacum]
Length = 259
Score = 362 bits (930), Expect = 4e-99
Identities = 187/255 (73%), Positives = 217/255 (84%), Gaps = 11/255 (4%)
Frame = -3
Query: 966 MLLRAASSFSLLNSNGGDHLLPPLFSSS-------VSPKT----FLPCATKGSNNRPLTG 820
MLL+ A +F+LLNS+G ++L P L +SS S K+ + CA+KGSN +PLTG
Sbjct: 1 MLLKLAPAFTLLNSHG-ENLSPMLSTSSQGFVLKNFSTKSRNGLLVVCASKGSNTKPLTG 59
Query: 819 VVFEPFEEVKKELDLVPTVPQASLARQKFVDECEAAINEQINVEYTVSYVYHAMFAYFDR 640
VVFEPFEEVKKEL LVPTVPQ SLAR K+ D+CEAA+NEQINVEY VSYVYH M+AYFDR
Sbjct: 60 VVFEPFEEVKKELMLVPTVPQVSLARHKYSDQCEAAVNEQINVEYNVSYVYHGMYAYFDR 119
Query: 639 DNVALKGLAKFFKESSMEEREHAEKLMEYQNKRGGRVKLQSIVMPPSEFEHEEKGDALYA 460
DNVALKGLA+FFKESS EER HAEKLMEYQNKRGG+VKLQSI+MP SEF+H E+GDALYA
Sbjct: 120 DNVALKGLARFFKESSEEERGHAEKLMEYQNKRGGKVKLQSILMPLSEFDHAEEGDALYA 179
Query: 459 MELALSLEKLTNEKLLNLHSVASKNNDAQLTDFIESEYLGEQVESIKPISEYVAQLRRVG 280
MELALSL KLTN+KLLNLH+VA++NND QL DF+ES+YL EQVE+IK ISEYVAQLRRVG
Sbjct: 180 MELALSLAKLTNQKLLNLHAVATRNNDVQLADFVESKYLREQVEAIKMISEYVAQLRRVG 239
Query: 279 QGHGTWHFNQMLLQD 235
+GHG WHF+QMLLQ+
Sbjct: 240 KGHGVWHFDQMLLQE 254
>gb|AAM11429.1| ferritin [Nicotiana tabacum]
Length = 251
Score = 362 bits (928), Expect = 7e-99
Identities = 187/253 (73%), Positives = 215/253 (84%), Gaps = 5/253 (1%)
Frame = -3
Query: 966 MLLRAASSFSLLNSNGGDHLLPPLFSSS--VSPKT---FLPCATKGSNNRPLTGVVFEPF 802
MLL+AA +F+LLN+ G + L PLFSSS SPK F+ A+K +N++PLTGVVFEPF
Sbjct: 1 MLLKAAPAFALLNTQGEN--LSPLFSSSKSFSPKNGNRFVVSASKATNHKPLTGVVFEPF 58
Query: 801 EEVKKELDLVPTVPQASLARQKFVDECEAAINEQINVEYTVSYVYHAMFAYFDRDNVALK 622
EE+KKEL LVP VP SL RQK+ D+CEAAINEQINVEY SYVYHAMFAYFDRDNVALK
Sbjct: 59 EELKKELMLVPAVPDTSLCRQKYSDDCEAAINEQINVEYNNSYVYHAMFAYFDRDNVALK 118
Query: 621 GLAKFFKESSMEEREHAEKLMEYQNKRGGRVKLQSIVMPPSEFEHEEKGDALYAMELALS 442
GLAKFFKESS+EEREHAEKLME+QNKRGGRVKL SI PP+EF+H EKGDALYAMELAL
Sbjct: 119 GLAKFFKESSLEEREHAEKLMEFQNKRGGRVKLLSICAPPTEFDHCEKGDALYAMELALC 178
Query: 441 LEKLTNEKLLNLHSVASKNNDAQLTDFIESEYLGEQVESIKPISEYVAQLRRVGQGHGTW 262
LEKLTN++LLNLH+VAS++ND L DF+ESE+L EQV++IK ISEYVAQLRRVGQGHG W
Sbjct: 179 LEKLTNQRLLNLHAVASRSNDVHLADFLESEFLVEQVDAIKKISEYVAQLRRVGQGHGVW 238
Query: 261 HFNQMLLQDGVAA 223
F+QMLL +G AA
Sbjct: 239 QFDQMLLNEGAAA 251
>dbj|BAB64537.1| ferritin [Glycine max]
Length = 247
Score = 356 bits (914), Expect = 3e-97
Identities = 189/250 (75%), Positives = 211/250 (83%), Gaps = 5/250 (2%)
Frame = -3
Query: 966 MLLR----AASSFSLLNSNGGDHLLPPLFSSSVSPKTFLPCATKGS-NNRPLTGVVFEPF 802
MLLR +ASS SL + N PP SV + + A KGS N+R LTGV+FEPF
Sbjct: 1 MLLRTAAASASSLSLFSPNAE----PP---RSVPARGLVVRAAKGSTNHRALTGVIFEPF 53
Query: 801 EEVKKELDLVPTVPQASLARQKFVDECEAAINEQINVEYTVSYVYHAMFAYFDRDNVALK 622
EEVKKELDLVPTVPQASLARQK+VDE E+A+NEQINVEY VSYVYHAMFAYFDRDNVAL+
Sbjct: 54 EEVKKELDLVPTVPQASLARQKYVDESESAVNEQINVEYNVSYVYHAMFAYFDRDNVALR 113
Query: 621 GLAKFFKESSMEEREHAEKLMEYQNKRGGRVKLQSIVMPPSEFEHEEKGDALYAMELALS 442
GLAKFFKESS EEREHAEKLMEYQNKRGG+VKLQSIVMP S+F+H +KGDAL+AMELALS
Sbjct: 114 GLAKFFKESSEEEREHAEKLMEYQNKRGGKVKLQSIVMPLSDFDHADKGDALHAMELALS 173
Query: 441 LEKLTNEKLLNLHSVASKNNDAQLTDFIESEYLGEQVESIKPISEYVAQLRRVGQGHGTW 262
LEKLTNEKLLNLHSVA+KN D QL DF+E+EYLGEQVE+IK ISEYVAQLR+ QGHG W
Sbjct: 174 LEKLTNEKLLNLHSVATKNGDVQLADFVETEYLGEQVEAIKRISEYVAQLRKSWQGHGVW 233
Query: 261 HFNQMLLQDG 232
HF+QMLL +G
Sbjct: 234 HFDQMLLHEG 243
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 910,687,110
Number of Sequences: 1393205
Number of extensions: 21338578
Number of successful extensions: 83585
Number of sequences better than 10.0: 401
Number of HSP's better than 10.0 without gapping: 70932
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 81559
length of database: 448,689,247
effective HSP length: 124
effective length of database: 275,931,827
effective search space used: 61532797421
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)