Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC015645A_C01 KMC015645A_c01
(560 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
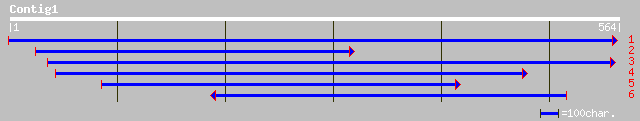
gb|AAF70243.1|AF213968_1 S-adenosyl-L-methionine Mg-protoporphyr... 108 6e-23
ref|NP_194238.1| magnesium-protoporphyrin IX methyltransferase -... 102 4e-21
dbj|BAA84812.1| Similar to magnesium-protoporphyrin IX methyltra... 92 6e-18
ref|NP_681241.1| Mg-protoporphyrin IX methyl transferase [Thermo... 70 1e-11
gb|ZP_00112151.1| hypothetical protein [Nostoc punctiforme] 68 9e-11
>gb|AAF70243.1|AF213968_1 S-adenosyl-L-methionine Mg-protoporphyrin IX methyltranserase
[Nicotiana tabacum]
Length = 325
Score = 108 bits (269), Expect = 6e-23
Identities = 51/55 (92%), Positives = 53/55 (95%)
Frame = -3
Query: 558 KRIGELFPGPSKATRAYLHSEGDVERALQKVGWKIRKRGLIATQFYFAKLVEAVP 394
KRIGELFPGPSKATRAYLH+E DVERALQK GWKIRKRGLIATQFYFAKL+EAVP
Sbjct: 270 KRIGELFPGPSKATRAYLHAEADVERALQKAGWKIRKRGLIATQFYFAKLIEAVP 324
>ref|NP_194238.1| magnesium-protoporphyrin IX methyltransferase - like protein;
protein id: At4g25080.1, supported by cDNA: gi_14334513,
supported by cDNA: gi_17104552 [Arabidopsis thaliana]
gi|7487951|pir||T05529 magnesium-protoporphyrin IX
methyltransferase (EC 2.1.1.-) - Arabidopsis thaliana
gi|4455251|emb|CAB36750.1| magnesium-protoporphyrin IX
methyltransferase-like protein [Arabidopsis thaliana]
gi|7269358|emb|CAB79417.1| magnesium-protoporphyrin IX
methyltransferase-like protein [Arabidopsis thaliana]
gi|14334514|gb|AAK59454.1| putative
magnesium-protoporphyrin IX methyltransferase
[Arabidopsis thaliana] gi|17104553|gb|AAL34165.1|
putative magnesium-protoporphyrin IX methyltransferase
[Arabidopsis thaliana]
Length = 312
Score = 102 bits (253), Expect = 4e-21
Identities = 48/56 (85%), Positives = 51/56 (90%)
Frame = -3
Query: 558 KRIGELFPGPSKATRAYLHSEGDVERALQKVGWKIRKRGLIATQFYFAKLVEAVPM 391
KRIGELFPGPSKATRAYLHSE DVERAL KVGWKI KRGL TQFYF++L+EAVPM
Sbjct: 257 KRIGELFPGPSKATRAYLHSEADVERALGKVGWKISKRGLTTTQFYFSRLIEAVPM 312
>dbj|BAA84812.1| Similar to magnesium-protoporphyrin IX methyltransferase (AL035523)
[Oryza sativa (japonica cultivar-group)]
gi|11875161|dbj|BAB19374.1| putative
magnesium-protoporphyrin IX methyltransferase [Oryza
sativa (japonica cultivar-group)]
gi|28190668|gb|AAO33146.1| putative
magnesium-protoporphyrin IX methyltransferase [Oryza
sativa (japonica cultivar-group)]
Length = 326
Score = 91.7 bits (226), Expect = 6e-18
Identities = 41/56 (73%), Positives = 48/56 (85%)
Frame = -3
Query: 558 KRIGELFPGPSKATRAYLHSEGDVERALQKVGWKIRKRGLIATQFYFAKLVEAVPM 391
KR+GELFPGPSKATRAYLHSE D+E AL+ GW++ RG I+TQFYFAKL EAVP+
Sbjct: 266 KRVGELFPGPSKATRAYLHSERDIEDALRDAGWRVANRGFISTQFYFAKLFEAVPI 321
>ref|NP_681241.1| Mg-protoporphyrin IX methyl transferase [Thermosynechococcus
elongatus BP-1] gi|22294172|dbj|BAC08003.1|
Mg-protoporphyrin IX methyl transferase
[Thermosynechococcus elongatus BP-1]
Length = 238
Score = 70.5 bits (171), Expect = 1e-11
Identities = 31/55 (56%), Positives = 41/55 (74%)
Frame = -3
Query: 558 KRIGELFPGPSKATRAYLHSEGDVERALQKVGWKIRKRGLIATQFYFAKLVEAVP 394
K+IGE FPG SKATRAYLHSE + L +GW+I + + T+FYF++L+EAVP
Sbjct: 182 KKIGEFFPGASKATRAYLHSEEFIVEILGNLGWRIERNAMTKTRFYFSRLLEAVP 236
>gb|ZP_00112151.1| hypothetical protein [Nostoc punctiforme]
Length = 228
Score = 67.8 bits (164), Expect = 9e-11
Identities = 29/53 (54%), Positives = 41/53 (76%)
Frame = -3
Query: 558 KRIGELFPGPSKATRAYLHSEGDVERALQKVGWKIRKRGLIATQFYFAKLVEA 400
K+IG FPGPSKATRAYLH E DV R L+ G+ ++++ + T+FYF++L+EA
Sbjct: 173 KKIGSFFPGPSKATRAYLHREADVVRILESNGFSLQRKAMTKTRFYFSRLLEA 225
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 474,794,337
Number of Sequences: 1393205
Number of extensions: 10255524
Number of successful extensions: 23454
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 22830
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 23450
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20095422690
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)