Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC015643A_C01 KMC015643A_c01
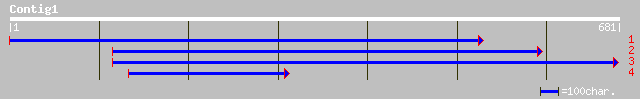
(627 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_190735.1| putative protein; protein id: At3g51670.1, supp... 264 6e-70
emb|CAB43521.1| hypothetical protein [Arabidopsis thaliana] 239 2e-62
ref|NP_564360.1| expressed protein; protein id: At1g30690.1, sup... 139 2e-32
gb|AAK59666.1| unknown protein [Arabidopsis thaliana] 139 2e-32
dbj|BAB86220.1| contains ESTs AU093915(E1276),AU162319(E60301)~s... 139 3e-32
>ref|NP_190735.1| putative protein; protein id: At3g51670.1, supported by cDNA:
gi_16930482 [Arabidopsis thaliana]
gi|11358091|pir||T46063 hypothetical protein T18N14.50 -
Arabidopsis thaliana gi|6580149|emb|CAB63153.1| putative
protein [Arabidopsis thaliana]
gi|16930483|gb|AAL31927.1|AF419595_1 AT3g51670/T18N14_50
[Arabidopsis thaliana] gi|25141223|gb|AAN73306.1|
At3g51670/T18N14_50 [Arabidopsis thaliana]
Length = 409
Score = 264 bits (675), Expect = 6e-70
Identities = 128/158 (81%), Positives = 145/158 (91%)
Frame = -3
Query: 625 SPFLTQRTKSKFVISKEGNAAETLYKFVRPEEIPVRYGGLCRPSDLQNGPPKPASEFTVK 446
SPFLTQRTKSKFV+SKEGNAAETLYKF+RPE+IPV+YGGL RP+D QNGPPKPASEF++K
Sbjct: 250 SPFLTQRTKSKFVMSKEGNAAETLYKFIRPEDIPVQYGGLSRPTDSQNGPPKPASEFSIK 309
Query: 445 GGEKVNIQIEGIEYGATITWDIVVGGWDLEYSAEFVPNAEGSYTIAVEKARKIEASEEAI 266
GGEKVNIQIEGIE GATITWDIVVGGWDLEYSAEFVPNAE SY I VEK +K++A++EA+
Sbjct: 310 GGEKVNIQIEGIEGGATITWDIVVGGWDLEYSAEFVPNAEESYAIVVEKPKKMKATDEAV 369
Query: 265 HNSFTLKEAGKMVLSVDNSASRKKKVAAYRYIVRKYST 152
NSFT EAGK++LSVDN+ SRKKKVAAYRY VRK +T
Sbjct: 370 CNSFTTVEAGKLILSVDNTLSRKKKVAAYRYTVRKSTT 407
>emb|CAB43521.1| hypothetical protein [Arabidopsis thaliana]
Length = 147
Score = 239 bits (610), Expect = 2e-62
Identities = 115/145 (79%), Positives = 132/145 (90%)
Frame = -3
Query: 586 ISKEGNAAETLYKFVRPEEIPVRYGGLCRPSDLQNGPPKPASEFTVKGGEKVNIQIEGIE 407
+SKEGNAAETLYKF+RPE+IPV+YGGL RP+D QNGPPKPASEF++KGGEKVNIQIEGIE
Sbjct: 1 MSKEGNAAETLYKFIRPEDIPVQYGGLSRPTDSQNGPPKPASEFSIKGGEKVNIQIEGIE 60
Query: 406 YGATITWDIVVGGWDLEYSAEFVPNAEGSYTIAVEKARKIEASEEAIHNSFTLKEAGKMV 227
GATITWDIVVGGWDLEYSAEFVPNAE SY I VEK +K++A++EA+ NSFT EAGK++
Sbjct: 61 GGATITWDIVVGGWDLEYSAEFVPNAEESYAIVVEKPKKMKATDEAVCNSFTTVEAGKLI 120
Query: 226 LSVDNSASRKKKVAAYRYIVRKYST 152
LSVDN+ SRKKKVAAYRY VRK +T
Sbjct: 121 LSVDNTLSRKKKVAAYRYTVRKSTT 145
>ref|NP_564360.1| expressed protein; protein id: At1g30690.1, supported by cDNA:
gi_14334977 [Arabidopsis thaliana]
gi|25518522|pir||D86432 hypothetical protein T5I8.14 -
Arabidopsis thaliana
gi|4587525|gb|AAD25756.1|AC007060_14 Contains the
PF|00650 CRAL/TRIO phosphatidyl-inositol-transfer
protein domain. ESTs gb|T76582, gb|N06574 and gb|Z25700
come from this gene. [Arabidopsis thaliana]
gi|24030399|gb|AAN41359.1| unknown protein [Arabidopsis
thaliana]
Length = 540
Score = 139 bits (351), Expect = 2e-32
Identities = 73/158 (46%), Positives = 100/158 (63%)
Frame = -3
Query: 625 SPFLTQRTKSKFVISKEGNAAETLYKFVRPEEIPVRYGGLCRPSDLQNGPPKPASEFTVK 446
SPFLTQRTKSKFV+++ ETL K++ +E+PV+YGG D + + SE VK
Sbjct: 384 SPFLTQRTKSKFVVARPAKVRETLLKYIPADELPVQYGGFKTVDDTEFSN-ETVSEVVVK 442
Query: 445 GGEKVNIQIEGIEYGATITWDIVVGGWDLEYSAEFVPNAEGSYTIAVEKARKIEASEEAI 266
G I+I E T+ WDI V GW++ Y EFVP EG+YT+ V+K +K+ A+E I
Sbjct: 443 PGSSETIEIPAPETEGTLVWDIAVLGWEVNYKEEFVPTEEGAYTVIVQKVKKMGANEGPI 502
Query: 265 HNSFTLKEAGKMVLSVDNSASRKKKVAAYRYIVRKYST 152
NSF +AGK+VL+VDN + +KKKV YRY + S+
Sbjct: 503 RNSFKNSQAGKIVLTVDNVSGKKKKV-LYRYRTKTESS 539
>gb|AAK59666.1| unknown protein [Arabidopsis thaliana]
Length = 540
Score = 139 bits (351), Expect = 2e-32
Identities = 73/158 (46%), Positives = 100/158 (63%)
Frame = -3
Query: 625 SPFLTQRTKSKFVISKEGNAAETLYKFVRPEEIPVRYGGLCRPSDLQNGPPKPASEFTVK 446
SPFLTQRTKSKFV+++ ETL K++ +E+PV+YGG D + + SE VK
Sbjct: 384 SPFLTQRTKSKFVVARPAKVRETLLKYIPADELPVQYGGFKTVDDTEFSN-ETVSEVVVK 442
Query: 445 GGEKVNIQIEGIEYGATITWDIVVGGWDLEYSAEFVPNAEGSYTIAVEKARKIEASEEAI 266
G I+I E T+ WDI V GW++ Y EFVP EG+YT+ V+K +K+ A+E I
Sbjct: 443 PGSSETIEIPAPETEGTLVWDIAVLGWEVNYKEEFVPTEEGAYTVIVQKVKKMGANEGPI 502
Query: 265 HNSFTLKEAGKMVLSVDNSASRKKKVAAYRYIVRKYST 152
NSF +AGK+VL+VDN + +KKKV YRY + S+
Sbjct: 503 RNSFKNSQAGKIVLTVDNVSGKKKKV-LYRYRTKTESS 539
>dbj|BAB86220.1| contains ESTs AU093915(E1276),AU162319(E60301)~similar to cytosolic
factor~unknown protein [Oryza sativa (japonica
cultivar-group)] gi|20804751|dbj|BAB92436.1| P0698A10.6
[Oryza sativa (japonica cultivar-group)]
Length = 613
Score = 139 bits (350), Expect = 3e-32
Identities = 71/158 (44%), Positives = 106/158 (66%), Gaps = 1/158 (0%)
Frame = -3
Query: 625 SPFLTQRTKSKFVISKEGNAAETLYKFVRPEEIPVRYGGLCRPSDLQNGPPKPASEFTVK 446
SPFLTQRTKSKF+ + +AETL++++ PE++PV++GGL + D + +E T+K
Sbjct: 455 SPFLTQRTKSKFIFASPAKSAETLFRYIAPEQVPVQFGGLFKEDDPEFTTSDAVTELTIK 514
Query: 445 GGEKVNIQIEGIEYGATITWDIVVGGWDLEYSAEFVPNAEGSYTIAVEKARKIEASEEAI 266
K ++I E +TI W++ V GW++ Y AEF P+AEG YT+ V+K RK+ A+EE I
Sbjct: 515 PSSKETVEIPVTE-NSTIGWELRVLGWEVSYGAEFTPDAEGGYTVIVQKTRKVPANEEPI 573
Query: 265 -HNSFTLKEAGKMVLSVDNSASRKKKVAAYRYIVRKYS 155
SF + E GK+VL+++N AS+KKK+ YR V+ S
Sbjct: 574 MKGSFKVGEPGKIVLTINNPASKKKKL-LYRSKVKSTS 610
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 553,534,579
Number of Sequences: 1393205
Number of extensions: 11979392
Number of successful extensions: 34437
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 33094
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 34403
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25586195130
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)