Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC015629A_C01 KMC015629A_c01
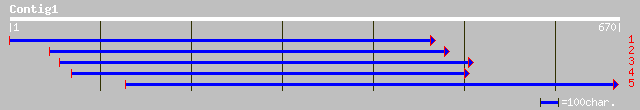
(669 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_568060.1| mitochondrial carrier protein family; protein i... 219 3e-56
ref|NP_564436.1| allinase, putative; protein id: At1g34065.1 [Ar... 201 7e-51
pir||T09362 hypothetical protein F23K16.90 - Arabidopsis thalian... 157 9e-38
ref|NP_014395.1| Member of family of mitochondrial carrier prote... 92 8e-18
emb|CAA07008.1| AgPET8 [Eremothecium gossypii] 91 1e-17
>ref|NP_568060.1| mitochondrial carrier protein family; protein id: At4g39460.1,
supported by cDNA: 32654., supported by cDNA:
gi_15028274, supported by cDNA: gi_19310698 [Arabidopsis
thaliana] gi|15028275|gb|AAK76726.1| putative
mitochondrial carrier protein [Arabidopsis thaliana]
gi|19310699|gb|AAL85080.1| putative mitochondrial
carrier protein [Arabidopsis thaliana]
Length = 325
Score = 219 bits (557), Expect = 3e-56
Identities = 108/134 (80%), Positives = 119/134 (88%)
Frame = -3
Query: 667 RSFLLRDLPFDAIQFCIYEQIRLGYMLAARRNLNDPENAVIGAFAGALTGAITTPLDVIK 488
RSFLLRDLPFDAIQFCIYEQ+ LGY AARR L+DPENA+IGAFAGALTGA+TTPLDVIK
Sbjct: 192 RSFLLRDLPFDAIQFCIYEQLCLGYKKAARRELSDPENALIGAFAGALTGAVTTPLDVIK 251
Query: 487 TRLMVQGSANQYKGIVDCVQTIMKEEGPRAFLKGIGPRVLWIGIGGSIFFGVLESSKRFL 308
TRLMVQGSA QY+GIVDCVQTI++EEG A LKGIGPRVLWIGIGGSIFFGVLES+KR L
Sbjct: 252 TRLMVQGSAKQYQGIVDCVQTIVREEGAPALLKGIGPRVLWIGIGGSIFFGVLESTKRTL 311
Query: 307 VERRPTLAQHSKSE 266
+RRP + +K E
Sbjct: 312 AQRRPNTVKETKEE 325
>ref|NP_564436.1| allinase, putative; protein id: At1g34065.1 [Arabidopsis thaliana]
Length = 327
Score = 201 bits (511), Expect = 7e-51
Identities = 94/122 (77%), Positives = 110/122 (90%)
Frame = -3
Query: 664 SFLLRDLPFDAIQFCIYEQIRLGYMLAARRNLNDPENAVIGAFAGALTGAITTPLDVIKT 485
SFLLRDLPFDA+QFC+YEQ+R+GY LAARR+LNDPENA+IGAFAGA+TG +TTPLDVIKT
Sbjct: 199 SFLLRDLPFDALQFCVYEQLRIGYKLAARRDLNDPENAMIGAFAGAVTGVLTTPLDVIKT 258
Query: 484 RLMVQGSANQYKGIVDCVQTIMKEEGPRAFLKGIGPRVLWIGIGGSIFFGVLESSKRFLV 305
RLMVQGS QYKG+ DC++TI++EEG A KG+GPRVLWIGIGGSIFFGVLE +K+ L
Sbjct: 259 RLMVQGSGTQYKGVSDCIKTIIREEGSSALWKGMGPRVLWIGIGGSIFFGVLEKTKQILS 318
Query: 304 ER 299
ER
Sbjct: 319 ER 320
Score = 40.0 bits (92), Expect = 0.029
Identities = 23/89 (25%), Positives = 41/89 (45%)
Frame = -3
Query: 559 ENAVIGAFAGALTGAITTPLDVIKTRLMVQGSANQYKGIVDCVQTIMKEEGPRAFLKGIG 380
E+ + G AG + A P+D IKTR+ V + G ++ G+G
Sbjct: 56 ESLITGGLAGVVVEAALYPIDTIKTRIQVVSPQTNFSG--------WWKDNMEGLYSGLG 107
Query: 379 PRVLWIGIGGSIFFGVLESSKRFLVERRP 293
++ + ++FFGV E +K+ L++ P
Sbjct: 108 GNLVGVLPASALFFGVYEPTKQKLLKVLP 136
Score = 34.7 bits (78), Expect = 1.2
Identities = 29/97 (29%), Positives = 45/97 (45%), Gaps = 4/97 (4%)
Frame = -3
Query: 646 LPFDAIQFCIYEQIRLGYMLAARRNLNDPENAVIGAFAGALTGAITT----PLDVIKTRL 479
LP A+ F +YE + + NL+ AV AGAL GA+++ P +V+K R+
Sbjct: 114 LPASALFFGVYEPTKQKLLKVLPDNLS----AVAHLAAGALGGAVSSIVRVPTEVVKQRM 169
Query: 478 MVQGSANQYKGIVDCVQTIMKEEGPRAFLKGIGPRVL 368
Q+ D V+ I+ +EG G G +L
Sbjct: 170 ----QTGQFVSAPDAVRLIIAKEGFGGMYAGYGSFLL 202
>pir||T09362 hypothetical protein F23K16.90 - Arabidopsis thaliana
gi|5042162|emb|CAB44681.1| mitochondrial carrier-like
protein [Arabidopsis thaliana]
gi|7270930|emb|CAB80609.1| mitochondrial carrier-like
protein [Arabidopsis thaliana]
Length = 330
Score = 157 bits (398), Expect = 9e-38
Identities = 78/93 (83%), Positives = 85/93 (90%)
Frame = -3
Query: 667 RSFLLRDLPFDAIQFCIYEQIRLGYMLAARRNLNDPENAVIGAFAGALTGAITTPLDVIK 488
RSFLLRDLPFDAIQFCIYEQ+ LGY AARR L+DPENA+IGAFAGALTGA+TTPLDVIK
Sbjct: 192 RSFLLRDLPFDAIQFCIYEQLCLGYKKAARRELSDPENALIGAFAGALTGAVTTPLDVIK 251
Query: 487 TRLMVQGSANQYKGIVDCVQTIMKEEGPRAFLK 389
TRLMVQGSA QY+GIVDCVQTI++EEG A LK
Sbjct: 252 TRLMVQGSAKQYQGIVDCVQTIVREEGAPALLK 284
>ref|NP_014395.1| Member of family of mitochondrial carrier proteins; Pet8p
[Saccharomyces cerevisiae]
gi|730302|sp|P38921|PET8_YEAST PUTATIVE MITOCHONDRIAL
CARRIER PROTEIN PET8 gi|630175|pir||S45458 PET8 protein
- yeast (Saccharomyces cerevisiae)
gi|495307|gb|AAA64802.1| Pet8p gi|496713|emb|CAA54377.1|
ORF N2012 [Saccharomyces cerevisiae]
gi|1301816|emb|CAA95862.1| ORF YNL003c [Saccharomyces
cerevisiae]
Length = 284
Score = 91.7 bits (226), Expect = 8e-18
Identities = 46/127 (36%), Positives = 74/127 (58%), Gaps = 1/127 (0%)
Frame = -3
Query: 658 LLRDLPFDAIQFCIYEQIRLGYMLAARRNLNDP-ENAVIGAFAGALTGAITTPLDVIKTR 482
++R++PF IQF +YE ++ + A ++ +P + A+ G+ AG + A TTPLD +KTR
Sbjct: 158 IMREIPFTCIQFPLYEYLKKTWAKANGQSQVEPWKGAICGSIAGGIAAATTTPLDFLKTR 217
Query: 481 LMVQGSANQYKGIVDCVQTIMKEEGPRAFLKGIGPRVLWIGIGGSIFFGVLESSKRFLVE 302
LM+ + ++ I +EEGP F G+GPR +WI GG+IF G+ E+ L +
Sbjct: 218 LMLNKTTASLGSVII---RIYREEGPAVFFSGVGPRTMWISAGGAIFLGMYETVHSLLSK 274
Query: 301 RRPTLAQ 281
PT +
Sbjct: 275 SFPTAGE 281
Score = 33.9 bits (76), Expect = 2.0
Identities = 25/88 (28%), Positives = 38/88 (42%)
Frame = -3
Query: 544 GAFAGALTGAITTPLDVIKTRLMVQGSANQYKGIVDCVQTIMKEEGPRAFLKGIGPRVLW 365
GA AG T + P+D IKTRL +G G + +G+G V+
Sbjct: 11 GAAAGTSTDLVFFPIDTIKTRLQAKGG-------------FFANGGYKGIYRGLGSAVVA 57
Query: 364 IGIGGSIFFGVLESSKRFLVERRPTLAQ 281
G S+FF + K V+ RP +++
Sbjct: 58 SAPGASLFFISYDYMK---VKSRPYISK 82
>emb|CAA07008.1| AgPET8 [Eremothecium gossypii]
Length = 271
Score = 90.9 bits (224), Expect = 1e-17
Identities = 45/113 (39%), Positives = 71/113 (62%), Gaps = 1/113 (0%)
Frame = -3
Query: 658 LLRDLPFDAIQFCIYEQIRLGYMLAAR-RNLNDPENAVIGAFAGALTGAITTPLDVIKTR 482
++R++PF IQF +YE ++ + A ++ + AV G+ AG + A TTPLDV+KTR
Sbjct: 157 IMREIPFTCIQFPLYEYLKKKWAAYAEIERVSAWQGAVCGSLAGGIAAAATTPLDVLKTR 216
Query: 481 LMVQGSANQYKGIVDCVQTIMKEEGPRAFLKGIGPRVLWIGIGGSIFFGVLES 323
+M+ + ++ +T+ +EEG R F +GIGPR +WI GG+IF GV E+
Sbjct: 217 MMLH---ERRVPMLHLARTLFREEGARVFFRGIGPRTMWISAGGAIFLGVYEA 266
Score = 41.6 bits (96), Expect = 0.010
Identities = 29/96 (30%), Positives = 43/96 (44%)
Frame = -3
Query: 550 VIGAFAGALTGAITTPLDVIKTRLMVQGSANQYKGIVDCVQTIMKEEGPRAFLKGIGPRV 371
V GA AG T + P+D +KTRL +G G R +G+G V
Sbjct: 10 VSGAAAGTSTDVVFFPIDTLKTRLQAKGG-------------FFHNGGYRGIYRGLGSAV 56
Query: 370 LWIGIGGSIFFGVLESSKRFLVERRPTLAQHSKSER 263
+ G S+FF +S K+ L RP + + + SE+
Sbjct: 57 VASAPGASLFFVTYDSMKQQL---RPVMGRWTASEQ 89
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 559,522,543
Number of Sequences: 1393205
Number of extensions: 12392909
Number of successful extensions: 37002
Number of sequences better than 10.0: 911
Number of HSP's better than 10.0 without gapping: 32709
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 35916
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 29138478756
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)