Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC015565A_C01 KMC015565A_c01
(794 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
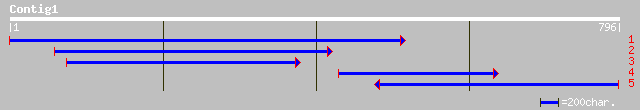
pir||T11855 DnaJ protein homolog - kidney bean (fragment) gi|168... 171 9e-60
ref|NP_179378.1| putative DnaJ protein; protein id: At2g17880.1,... 91 6e-28
ref|NP_195328.1| DnaJ-like protein; protein id: At4g36040.1 [Ara... 77 3e-24
emb|CAC03599.1| J11 protein [Arabidopsis thaliana] 77 3e-24
ref|NP_187939.1| DnaJ protein, putative; protein id: At3g13310.1... 49 9e-10
>pir||T11855 DnaJ protein homolog - kidney bean (fragment)
gi|1684851|gb|AAB36543.1| DnaJ-like protein [Phaseolus
vulgaris]
Length = 161
Score = 171 bits (433), Expect(2) = 9e-60
Identities = 92/119 (77%), Positives = 103/119 (86%), Gaps = 4/119 (3%)
Frame = -2
Query: 793 SLSAANLTGNTVATPPRRVRSRPVVSFATATATATSTA----DARSTWTEQPRPSSTYLI 626
SL A N +G +A+PP RVRSRP+V+FATATATAT+TA +ARS+WTE+PRPS YL
Sbjct: 5 SLPAVNFSGKVLASPPCRVRSRPIVAFATATATATATATSTEEARSSWTEKPRPS--YLN 62
Query: 625 SPCSSLYEILGIPAGASSQEIKAAYRRLARVCHHTDVAAIDRKNSSADDFMKIHAAYST 449
S CSSLY+ILGIPAGASSQEIKAAYRRLARVC H DVAAIDRKNSSAD+FMKIHAAYST
Sbjct: 63 SSCSSLYDILGIPAGASSQEIKAAYRRLARVC-HPDVAAIDRKNSSADEFMKIHAAYST 120
Score = 82.0 bits (201), Expect(2) = 9e-60
Identities = 35/41 (85%), Positives = 38/41 (92%)
Frame = -3
Query: 447 LSDPEKRATYDRSLFRRQRPMSTAVVFSGYSSRNWETDQCW 325
LSDP+KRA YDRSLFRRQRP+STA VFSGY+ RNWETDQCW
Sbjct: 121 LSDPDKRANYDRSLFRRQRPLSTAAVFSGYTRRNWETDQCW 161
>ref|NP_179378.1| putative DnaJ protein; protein id: At2g17880.1, supported by cDNA:
22711., supported by cDNA: gi_14190374, supported by
cDNA: gi_20147114 [Arabidopsis thaliana]
gi|7486985|pir||T00836 probable dnaJ protein At2g17880
[imported] - Arabidopsis thaliana
gi|14190375|gb|AAK55668.1|AF378865_1 At2g17880
[Arabidopsis thaliana] gi|20147115|gb|AAM10274.1|
At2g17880/At2g17880 [Arabidopsis thaliana]
gi|21592352|gb|AAM64303.1| putative DnaJ protein
[Arabidopsis thaliana]
Length = 160
Score = 90.5 bits (223), Expect(2) = 6e-28
Identities = 58/115 (50%), Positives = 72/115 (62%)
Frame = -2
Query: 793 SLSAANLTGNTVATPPRRVRSRPVVSFATATATATSTADARSTWTEQPRPSSTYLISPCS 614
S S+++ + +PP R+ SR S +T TA+ T D R S+T +
Sbjct: 15 SSSSSSSPSFSSTSPPSRI-SRISPSLSTTTASYTCAEDLPRLRQIPQRFSAT------A 67
Query: 613 SLYEILGIPAGASSQEIKAAYRRLARVCHHTDVAAIDRKNSSADDFMKIHAAYST 449
SLYEIL IP G++SQEIK+AYRRLAR+C H DVA R NSSADDFMKIHAAY T
Sbjct: 68 SLYEILEIPVGSTSQEIKSAYRRLARIC-HPDVARNSRDNSSADDFMKIHAAYCT 121
Score = 56.2 bits (134), Expect(2) = 6e-28
Identities = 24/41 (58%), Positives = 27/41 (65%)
Frame = -3
Query: 447 LSDPEKRATYDRSLFRRQRPMSTAVVFSGYSSRNWETDQCW 325
LSDPEKRA YDR R RP++ + Y RNWETDQCW
Sbjct: 122 LSDPEKRAVYDRRTLLRSRPLTAG--YGSYGGRNWETDQCW 160
>ref|NP_195328.1| DnaJ-like protein; protein id: At4g36040.1 [Arabidopsis thaliana]
gi|7487181|pir||T05496 hypothetical protein T19K4.170 -
Arabidopsis thaliana gi|3036808|emb|CAA18498.1|
DnaJ-like protein [Arabidopsis thaliana]
gi|7270556|emb|CAB81513.1| DnaJ-like protein
[Arabidopsis thaliana]
Length = 161
Score = 77.4 bits (189), Expect(2) = 3e-24
Identities = 48/105 (45%), Positives = 66/105 (62%), Gaps = 1/105 (0%)
Frame = -2
Query: 760 VATPPRRVRSRPVVSFATATATATSTADARSTWTEQPRPSSTYLISPCSSLYEILGIPAG 581
++ P R R P + A+ + T T + + PR +T +SLY++L +P G
Sbjct: 23 ISPPSRTARISPPLVSASCSYTYTEDSPRLH---QIPRRLTTVP----ASLYDVLEVPLG 75
Query: 580 ASSQEIKAAYRRLARVCHHTDVAAIDR-KNSSADDFMKIHAAYST 449
A+SQ+IK+AYRRLAR+C H DVA DR +SSAD+FMKIHAAY T
Sbjct: 76 ATSQDIKSAYRRLARIC-HPDVAGTDRTSSSSADEFMKIHAAYCT 119
Score = 57.0 bits (136), Expect(2) = 3e-24
Identities = 25/42 (59%), Positives = 29/42 (68%), Gaps = 1/42 (2%)
Frame = -3
Query: 447 LSDPEKRATYDRSLFRRQRPMSTAVVFSG-YSSRNWETDQCW 325
LSDPEKR+ YDR + RR RP++ G Y RNWETDQCW
Sbjct: 120 LSDPEKRSVYDRRMLRRSRPLTVGTSGLGSYVGRNWETDQCW 161
>emb|CAC03599.1| J11 protein [Arabidopsis thaliana]
Length = 161
Score = 77.4 bits (189), Expect(2) = 3e-24
Identities = 48/105 (45%), Positives = 66/105 (62%), Gaps = 1/105 (0%)
Frame = -2
Query: 760 VATPPRRVRSRPVVSFATATATATSTADARSTWTEQPRPSSTYLISPCSSLYEILGIPAG 581
++ P R R P + A+ + T T + + PR +T +SLY++L +P G
Sbjct: 23 ISPPSRTARISPPLVSASCSYTYTEDSPRLH---QIPRRLTTVP----ASLYDVLEVPLG 75
Query: 580 ASSQEIKAAYRRLARVCHHTDVAAIDR-KNSSADDFMKIHAAYST 449
A+SQ+IK+AYRRLAR+C H DVA DR +SSAD+FMKIHAAY T
Sbjct: 76 ATSQDIKSAYRRLARIC-HPDVAGTDRTSSSSADEFMKIHAAYCT 119
Score = 57.0 bits (136), Expect(2) = 3e-24
Identities = 25/42 (59%), Positives = 29/42 (68%), Gaps = 1/42 (2%)
Frame = -3
Query: 447 LSDPEKRATYDRSLFRRQRPMSTAVVFSG-YSSRNWETDQCW 325
LSDPEKR+ YDR + RR RP++ G Y RNWETDQCW
Sbjct: 120 LSDPEKRSVYDRRMLRRSRPLTVGTSGLGRYVGRNWETDQCW 161
>ref|NP_187939.1| DnaJ protein, putative; protein id: At3g13310.1, supported by cDNA:
31309. [Arabidopsis thaliana] gi|9294537|dbj|BAB02800.1|
DnaJ-like protein [Arabidopsis thaliana]
gi|21592683|gb|AAM64632.1| DnaJ protein, putative
[Arabidopsis thaliana]
Length = 157
Score = 48.5 bits (114), Expect(2) = 9e-10
Identities = 28/56 (50%), Positives = 33/56 (58%)
Frame = -2
Query: 616 SSLYEILGIPAGASSQEIKAAYRRLARVCHHTDVAAIDRKNSSADDFMKIHAAYST 449
SSLYE+L + AS EIK AYR LA+V H D S DFM+IH AY+T
Sbjct: 63 SSLYELLKVNETASLTEIKTAYRSLAKVYHP------DASESDGRDFMEIHKAYAT 112
Score = 37.0 bits (84), Expect(2) = 9e-10
Identities = 21/45 (46%), Positives = 28/45 (61%), Gaps = 4/45 (8%)
Frame = -3
Query: 447 LSDPEKRATYDRSL-FRRQRPMSTAVVFSGY---SSRNWETDQCW 325
L+DP RA YD +L R+R + A+ SG ++R WETDQCW
Sbjct: 113 LADPTTRAIYDSTLRVPRRRVHAGAMGRSGRVYATTRRWETDQCW 157
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 694,118,219
Number of Sequences: 1393205
Number of extensions: 17343728
Number of successful extensions: 217000
Number of sequences better than 10.0: 2031
Number of HSP's better than 10.0 without gapping: 82865
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 151063
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 40055936206
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)