Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC015545A_C01 KMC015545A_c01
(728 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
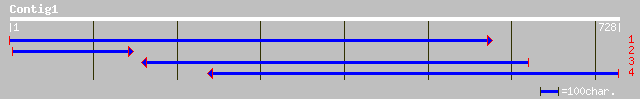
gb|AAK83088.1|AF290200_1 Pin1-type peptidyl-prolyl cis/trans iso... 201 7e-51
ref|NP_179395.1| putative peptidyl-prolyl cis-trans isomerase; p... 199 4e-50
pdb|1J6Y|A Chain A, Solution Structure Of Pin1at From Arabidopsi... 199 4e-50
emb|CAB94994.1| peptidyl-prolyl cis-trans isomerase [Digitalis l... 183 2e-45
gb|AAB36960.1| PinA [Dictyostelium discoideum] 131 1e-29
>gb|AAK83088.1|AF290200_1 Pin1-type peptidyl-prolyl cis/trans isomerase [Malus x domestica]
Length = 121
Score = 201 bits (512), Expect = 7e-51
Identities = 96/115 (83%), Positives = 109/115 (94%)
Frame = +3
Query: 123 GGEVRASHILIKHQGSRRKASWKDPEGRVIKNTTRESAVAQLSTLREDILSGKANFEDVA 302
G +VRASHILIKHQGSRRKASWKDPEG++I+NTTR+SAV+QL LR+DILSGKA F+D+A
Sbjct: 6 GNQVRASHILIKHQGSRRKASWKDPEGQIIRNTTRDSAVSQLKALRDDILSGKAKFDDLA 65
Query: 303 SRISDCSSAKRGGDLGPFGRGQMQKPFEEATFALKVGDISEIVDTDSGVHIIKRT 467
+R SDCSSAKRGGDLGPFGR QMQKPFEEATFALKVG++S+IVDTDSGVHIIKRT
Sbjct: 66 ARYSDCSSAKRGGDLGPFGRNQMQKPFEEATFALKVGEMSDIVDTDSGVHIIKRT 120
>ref|NP_179395.1| putative peptidyl-prolyl cis-trans isomerase; protein id:
At2g18040.1, supported by cDNA: 12602., supported by
cDNA: gi_13430811, supported by cDNA: gi_15810550,
supported by cDNA: gi_15982814 [Arabidopsis thaliana]
gi|25411826|pir||E84559 probable peptidyl-prolyl
cis-trans isomerase [imported] - Arabidopsis thaliana
gi|4406814|gb|AAD20122.1| putative peptidyl-prolyl
cis-trans isomerase [Arabidopsis thaliana]
gi|13430812|gb|AAK26028.1|AF360318_1 putative
peptidyl-prolyl cis-trans isomerase [Arabidopsis
thaliana] gi|15810551|gb|AAL07163.1| putative
peptidyl-prolyl cis-trans isomerase [Arabidopsis
thaliana] gi|15982815|gb|AAL09755.1| At2g18040/T27K22.9
[Arabidopsis thaliana] gi|21537274|gb|AAM61615.1|
putative peptidyl-prolyl cis-trans isomerase
[Arabidopsis thaliana]
Length = 119
Score = 199 bits (505), Expect = 4e-50
Identities = 95/113 (84%), Positives = 108/113 (95%)
Frame = +3
Query: 129 EVRASHILIKHQGSRRKASWKDPEGRVIKNTTRESAVAQLSTLREDILSGKANFEDVASR 308
+V+ASHILIKHQGSRRKASWKDPEG++I TTRE+AV QL ++REDI+SGKANFE+VA+R
Sbjct: 6 QVKASHILIKHQGSRRKASWKDPEGKIILTTTREAAVEQLKSIREDIVSGKANFEEVATR 65
Query: 309 ISDCSSAKRGGDLGPFGRGQMQKPFEEATFALKVGDISEIVDTDSGVHIIKRT 467
+SDCSSAKRGGDLG FGRGQMQKPFEEAT+ALKVGDIS+IVDTDSGVHIIKRT
Sbjct: 66 VSDCSSAKRGGDLGSFGRGQMQKPFEEATYALKVGDISDIVDTDSGVHIIKRT 118
>pdb|1J6Y|A Chain A, Solution Structure Of Pin1at From Arabidopsis Thaliana
Length = 139
Score = 199 bits (505), Expect = 4e-50
Identities = 95/113 (84%), Positives = 108/113 (95%)
Frame = +3
Query: 129 EVRASHILIKHQGSRRKASWKDPEGRVIKNTTRESAVAQLSTLREDILSGKANFEDVASR 308
+V+ASHILIKHQGSRRKASWKDPEG++I TTRE+AV QL ++REDI+SGKANFE+VA+R
Sbjct: 26 QVKASHILIKHQGSRRKASWKDPEGKIILTTTREAAVEQLKSIREDIVSGKANFEEVATR 85
Query: 309 ISDCSSAKRGGDLGPFGRGQMQKPFEEATFALKVGDISEIVDTDSGVHIIKRT 467
+SDCSSAKRGGDLG FGRGQMQKPFEEAT+ALKVGDIS+IVDTDSGVHIIKRT
Sbjct: 86 VSDCSSAKRGGDLGSFGRGQMQKPFEEATYALKVGDISDIVDTDSGVHIIKRT 138
>emb|CAB94994.1| peptidyl-prolyl cis-trans isomerase [Digitalis lanata]
Length = 118
Score = 183 bits (465), Expect = 2e-45
Identities = 89/113 (78%), Positives = 103/113 (90%)
Frame = +3
Query: 129 EVRASHILIKHQGSRRKASWKDPEGRVIKNTTRESAVAQLSTLREDILSGKANFEDVASR 308
+VRASHILIKHQGSRRK+SWKDP+G +I TTR+ AV+QL +LR+++LS A+F D+ASR
Sbjct: 5 KVRASHILIKHQGSRRKSSWKDPDGSLISATTRDDAVSQLQSLRQELLSDPASFSDLASR 64
Query: 309 ISDCSSAKRGGDLGPFGRGQMQKPFEEATFALKVGDISEIVDTDSGVHIIKRT 467
S CSSAKRGGDLGPFGRGQMQKPFEEATFALKVG+IS+IVDTDSGVHIIKRT
Sbjct: 65 HSHCSSAKRGGDLGPFGRGQMQKPFEEATFALKVGEISDIVDTDSGVHIIKRT 117
>gb|AAB36960.1| PinA [Dictyostelium discoideum]
Length = 243
Score = 131 bits (329), Expect = 1e-29
Identities = 65/111 (58%), Positives = 85/111 (76%)
Frame = +3
Query: 132 VRASHILIKHQGSRRKASWKDPEGRVIKNTTRESAVAQLSTLREDILSGKANFEDVASRI 311
V H+L+KHQGSR +SW+ E ++ + T+E A+A+L+ R I+SG A FED+A +
Sbjct: 135 VTCRHLLVKHQGSRNPSSWR--ESKITR--TKERAIAKLNEYRATIISGSATFEDLAHKN 190
Query: 312 SDCSSAKRGGDLGPFGRGQMQKPFEEATFALKVGDISEIVDTDSGVHIIKR 464
SDCSSAKRGG L PF RGQMQ+PFE+ F+LKVG++S IVDTDSGVHII+R
Sbjct: 191 SDCSSAKRGGYLDPFKRGQMQRPFEDCAFSLKVGEVSGIVDTDSGVHIIER 241
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 618,527,568
Number of Sequences: 1393205
Number of extensions: 13307572
Number of successful extensions: 45770
Number of sequences better than 10.0: 229
Number of HSP's better than 10.0 without gapping: 42915
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 45620
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 34343566934
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)