Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC015414A_C01 KMC015414A_c01
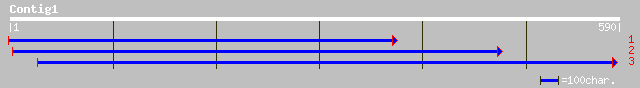
(590 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_563918.1| expressed protein; protein id: At1g12850.1, sup... 150 1e-35
gb|AAM63247.1| putative fructose-2,6-bisphosphatase [Arabidopsis... 149 2e-35
emb|CAC81923.1| putative Fructose-2,6-bisphosphatase [Silene lat... 143 2e-33
dbj|BAB01224.1| gene_id:MDJ14.7~similar to unknown protein~sp|P0... 142 3e-33
gb|AAM23298.1| X4 protein [Silene latifolia] 141 5e-33
>ref|NP_563918.1| expressed protein; protein id: At1g12850.1, supported by cDNA:
207684., supported by cDNA: gi_14517375 [Arabidopsis
thaliana] gi|14517376|gb|AAK62579.1| At1g12850/F13K23_8
[Arabidopsis thaliana] gi|21360535|gb|AAM47383.1|
At1g12850/F13K23_8 [Arabidopsis thaliana]
Length = 405
Score = 150 bits (379), Expect = 1e-35
Identities = 74/115 (64%), Positives = 87/115 (75%), Gaps = 1/115 (0%)
Frame = -1
Query: 569 HRPAFSRKKSGKSRLQFMTTTGDEVEDEVHSSNVNHQISLHNSSFSSFSPSVS-CIGLFT 393
HRP +RKKSGKSR Q M TGD E SN N + L + + S S +S CIG+FT
Sbjct: 291 HRPILTRKKSGKSRFQVMNATGDHEGSEEIFSNHNDEQHLGDINIKSSSSQLSTCIGVFT 350
Query: 392 HSVPIKCLLTGLLGCSPLMLHKFCIEDSSVTVLQHSVRTGWQIKRLNDTAHLRIL 228
HS+PIKCLLTG+LGCSP+M HK C+EDSSVTVLQHS +TGWQ+KRLNDTAHLR+L
Sbjct: 351 HSLPIKCLLTGILGCSPVMTHKICVEDSSVTVLQHSWKTGWQVKRLNDTAHLRLL 405
>gb|AAM63247.1| putative fructose-2,6-bisphosphatase [Arabidopsis thaliana]
Length = 405
Score = 149 bits (377), Expect = 2e-35
Identities = 74/115 (64%), Positives = 87/115 (75%), Gaps = 1/115 (0%)
Frame = -1
Query: 569 HRPAFSRKKSGKSRLQFMTTTGDEVEDEVHSSNVNHQISLHNSSFSSFSPSVS-CIGLFT 393
HRP +RKKSGKSR Q M TGD E SN N + L + + S S +S CIG+FT
Sbjct: 291 HRPILTRKKSGKSRFQVMNATGDHEGSEEIFSNHNDEQHLGDINIKSSSSLLSTCIGVFT 350
Query: 392 HSVPIKCLLTGLLGCSPLMLHKFCIEDSSVTVLQHSVRTGWQIKRLNDTAHLRIL 228
HS+PIKCLLTG+LGCSP+M HK C+EDSSVTVLQHS +TGWQ+KRLNDTAHLR+L
Sbjct: 351 HSLPIKCLLTGILGCSPVMTHKICVEDSSVTVLQHSWKTGWQVKRLNDTAHLRLL 405
>emb|CAC81923.1| putative Fructose-2,6-bisphosphatase [Silene latifolia]
Length = 426
Score = 143 bits (360), Expect = 2e-33
Identities = 74/122 (60%), Positives = 93/122 (75%), Gaps = 2/122 (1%)
Frame = -1
Query: 587 DSPNRHHRPAFSRKKSGKSRLQFMTTTG-DEVEDEVHSSNV-NHQISLHNSSFSSFSPSV 414
D RH + SRKKSGKSRLQ +TTTG D ED++ + + + S+ ++ S+S V
Sbjct: 307 DLLQRHRQ--LSRKKSGKSRLQVVTTTGVDYHEDDMSPKELPSCRSSISETNARSYSSGV 364
Query: 413 SCIGLFTHSVPIKCLLTGLLGCSPLMLHKFCIEDSSVTVLQHSVRTGWQIKRLNDTAHLR 234
+CIG+FTHSVPIKCLLTG+LGCSP + HK C++DSSVTVLQHS +TGWQIKRLND AHLR
Sbjct: 365 TCIGVFTHSVPIKCLLTGILGCSPAISHKLCVDDSSVTVLQHSWKTGWQIKRLNDVAHLR 424
Query: 233 IL 228
+L
Sbjct: 425 LL 426
>dbj|BAB01224.1| gene_id:MDJ14.7~similar to unknown protein~sp|P07953 [Arabidopsis
thaliana] gi|26449922|dbj|BAC42082.1| unknown protein
[Arabidopsis thaliana]
Length = 399
Score = 142 bits (358), Expect = 3e-33
Identities = 75/125 (60%), Positives = 91/125 (72%), Gaps = 4/125 (3%)
Frame = -1
Query: 590 WDSPNRHHRPAFSRKKSGKSRLQFMTT----TGDEVEDEVHSSNVNHQISLHNSSFSSFS 423
WD ++H RP+ +RKKSGKSRLQ MT G+ +EV N NH +SS S
Sbjct: 284 WDLLHKH-RPSLTRKKSGKSRLQVMTNHEPDDGNSPREEV---NHNHTDLSDSSSLIS-- 337
Query: 422 PSVSCIGLFTHSVPIKCLLTGLLGCSPLMLHKFCIEDSSVTVLQHSVRTGWQIKRLNDTA 243
+C+G+FTHS+PIKCLLTG+LGCSP+M HK C+EDSSVTVLQHS R GWQIKR+NDTA
Sbjct: 338 ---NCVGVFTHSLPIKCLLTGILGCSPVMTHKICVEDSSVTVLQHSWRNGWQIKRMNDTA 394
Query: 242 HLRIL 228
HLR+L
Sbjct: 395 HLRLL 399
>gb|AAM23298.1| X4 protein [Silene latifolia]
Length = 372
Score = 141 bits (356), Expect = 5e-33
Identities = 73/122 (59%), Positives = 90/122 (72%), Gaps = 2/122 (1%)
Frame = -1
Query: 587 DSPNRHHRPAFSRKKSGKSRLQFMTTTGDEVEDEVHSSNV--NHQISLHNSSFSSFSPSV 414
D RH + SRKKSGKSRLQ +TTTG ++ S + + S+ ++ SFS V
Sbjct: 253 DLLQRHRQ--LSRKKSGKSRLQVVTTTGVHYHEDDMSPKEPPSCRSSISETNARSFSSGV 310
Query: 413 SCIGLFTHSVPIKCLLTGLLGCSPLMLHKFCIEDSSVTVLQHSVRTGWQIKRLNDTAHLR 234
+CIG+FTHSVPIKCLLTG+LGCSP + HK C++DSSVTVLQHS +TGWQIKRLND AHLR
Sbjct: 311 TCIGVFTHSVPIKCLLTGILGCSPAISHKLCVDDSSVTVLQHSWKTGWQIKRLNDVAHLR 370
Query: 233 IL 228
+L
Sbjct: 371 LL 372
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 492,876,985
Number of Sequences: 1393205
Number of extensions: 10271446
Number of successful extensions: 24724
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 24057
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24702
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 22569056698
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)