Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC015368A_C01 KMC015368A_c01
(685 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
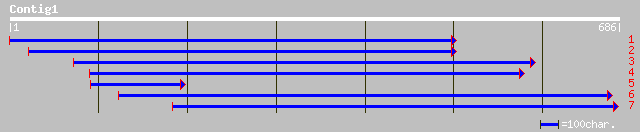
ref|NP_194697.1| nucleotide pyrophosphatase - like protein; prot... 192 3e-48
gb|AAK43863.1|AF370486_1 nucleotide pyrophosphatase-like protein... 192 3e-48
dbj|BAB20681.1| nucleotide pyrophosphatase-like protein [Spinaci... 190 2e-47
pir||T03293 probable phosphodiesterase I (EC 3.1.4.1) / nucleoti... 180 2e-44
ref|NP_194700.1| putative protein; protein id: At4g29710.1 [Arab... 167 2e-40
>ref|NP_194697.1| nucleotide pyrophosphatase - like protein; protein id: At4g29680.1,
supported by cDNA: gi_13877570 [Arabidopsis thaliana]
gi|7487979|pir||T09931 probable phosphodiesterase I (EC
3.1.4.1) / nucleotide diphosphatase (EC 3.6.1.9)
T16L4.190 - Arabidopsis thaliana
gi|5123562|emb|CAB45328.1| nucleotide
pyrophosphatase-like protein [Arabidopsis thaliana]
gi|7269867|emb|CAB79726.1| nucleotide
pyrophosphatase-like protein [Arabidopsis thaliana]
Length = 496
Score = 192 bits (489), Expect = 3e-48
Identities = 91/117 (77%), Positives = 100/117 (84%)
Frame = -1
Query: 685 GNVENGGKLKVYLKEDLPERLHYVASDRIPPIIGLVHEGYKVEQSRTNEKECAGAHGYDN 506
G VENG LKVYLKEDLP RLHYV SDRIPPIIGLV EG+KVEQ ++ KEC GAHGYDN
Sbjct: 377 GKVENGKYLKVYLKEDLPSRLHYVDSDRIPPIIGLVDEGFKVEQKKSKAKECGGAHGYDN 436
Query: 505 AFFSMRTIFVGHGPQFARGRKVPSFENVQIYNVVTSILNIKGAPNNGSTSFPESILL 335
AFFSMRTIF+GHGP F++GRKVPSFENVQIYNV++SIL +K APNNGS F SILL
Sbjct: 437 AFFSMRTIFIGHGPMFSKGRKVPSFENVQIYNVISSILGLKAAPNNGSDEFSSSILL 493
>gb|AAK43863.1|AF370486_1 nucleotide pyrophosphatase-like protein [Arabidopsis thaliana]
gi|23198336|gb|AAN15695.1| nucleotide
pyrophosphatase-like protein [Arabidopsis thaliana]
Length = 234
Score = 192 bits (489), Expect = 3e-48
Identities = 91/117 (77%), Positives = 100/117 (84%)
Frame = -1
Query: 685 GNVENGGKLKVYLKEDLPERLHYVASDRIPPIIGLVHEGYKVEQSRTNEKECAGAHGYDN 506
G VENG LKVYLKEDLP RLHYV SDRIPPIIGLV EG+KVEQ ++ KEC GAHGYDN
Sbjct: 115 GKVENGKYLKVYLKEDLPSRLHYVDSDRIPPIIGLVDEGFKVEQKKSKAKECGGAHGYDN 174
Query: 505 AFFSMRTIFVGHGPQFARGRKVPSFENVQIYNVVTSILNIKGAPNNGSTSFPESILL 335
AFFSMRTIF+GHGP F++GRKVPSFENVQIYNV++SIL +K APNNGS F SILL
Sbjct: 175 AFFSMRTIFIGHGPMFSKGRKVPSFENVQIYNVISSILGLKAAPNNGSDEFSSSILL 231
>dbj|BAB20681.1| nucleotide pyrophosphatase-like protein [Spinacia oleracea]
Length = 479
Score = 190 bits (482), Expect = 2e-47
Identities = 90/120 (75%), Positives = 102/120 (85%)
Frame = -1
Query: 685 GNVENGGKLKVYLKEDLPERLHYVASDRIPPIIGLVHEGYKVEQSRTNEKECAGAHGYDN 506
G V NG LKVYLKEDLP+RLHY S RIPPIIGL+ EGYKVEQ +N+ EC GAHGYDN
Sbjct: 360 GKVVNGKHLKVYLKEDLPKRLHYSDSYRIPPIIGLLDEGYKVEQKDSNKNECGGAHGYDN 419
Query: 505 AFFSMRTIFVGHGPQFARGRKVPSFENVQIYNVVTSILNIKGAPNNGSTSFPESILLSTA 326
+FSMRTIF+ HGPQFA+GRKVPSFENVQIYN+VTSIL+++GAPNNGS SFP S+LL A
Sbjct: 420 EYFSMRTIFIAHGPQFAKGRKVPSFENVQIYNLVTSILDVEGAPNNGSVSFPNSVLLPHA 479
>pir||T03293 probable phosphodiesterase I (EC 3.1.4.1) / nucleotide
diphosphatase (EC 3.6.1.9) - rice
gi|818849|gb|AAA67067.1| nucleotide pyrophosphatase
precursor gi|7638402|gb|AAF65458.1|AF245483_1 OSE4
[Oryza sativa] gi|14209591|dbj|BAB56086.1| nucleotide
pyrophosphatase homolog [Oryza sativa (japonica
cultivar-group)]
Length = 479
Score = 180 bits (456), Expect = 2e-44
Identities = 84/117 (71%), Positives = 98/117 (82%)
Frame = -1
Query: 685 GNVENGGKLKVYLKEDLPERLHYVASDRIPPIIGLVHEGYKVEQSRTNEKECAGAHGYDN 506
G VENG L++YLKEDLP RLHY S RIPPIIGL EGYKVE R+++ EC GAHGYDN
Sbjct: 360 GKVENGEYLRMYLKEDLPSRLHYADSYRIPPIIGLPEEGYKVEMKRSDKNECGGAHGYDN 419
Query: 505 AFFSMRTIFVGHGPQFARGRKVPSFENVQIYNVVTSILNIKGAPNNGSTSFPESILL 335
AFFSMRTIF+ HGP+F GR VPSFENV+IYNV+ SILN++ APNNGS+SFP++ILL
Sbjct: 420 AFFSMRTIFIAHGPRFEGGRVVPSFENVEIYNVIASILNLEPAPNNGSSSFPDTILL 476
>ref|NP_194700.1| putative protein; protein id: At4g29710.1 [Arabidopsis thaliana]
gi|7487098|pir||T09934 hypothetical protein T16L4.220 -
Arabidopsis thaliana gi|5123565|emb|CAB45331.1| putative
protein [Arabidopsis thaliana]
gi|7269870|emb|CAB79729.1| putative protein [Arabidopsis
thaliana]
Length = 133
Score = 167 bits (422), Expect = 2e-40
Identities = 82/117 (70%), Positives = 93/117 (79%)
Frame = -1
Query: 685 GNVENGGKLKVYLKEDLPERLHYVASDRIPPIIGLVHEGYKVEQSRTNEKECAGAHGYDN 506
G V+NG L VYLKE LP+RLHY S RIPPIIG+V EG V Q+RTN +EC G HGYDN
Sbjct: 8 GKVKNGEFLTVYLKEKLPDRLHYSQSYRIPPIIGMVGEGLIVRQNRTNAQECYGDHGYDN 67
Query: 505 AFFSMRTIFVGHGPQFARGRKVPSFENVQIYNVVTSILNIKGAPNNGSTSFPESILL 335
FFSMRTIFVGHG +F RG+KVPSFENVQIY+VV IL ++ APNNGS+ FP SILL
Sbjct: 68 KFFSMRTIFVGHGSRFRRGKKVPSFENVQIYSVVADILGLRPAPNNGSSLFPRSILL 124
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 596,535,080
Number of Sequences: 1393205
Number of extensions: 13314268
Number of successful extensions: 34675
Number of sequences better than 10.0: 106
Number of HSP's better than 10.0 without gapping: 31742
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 33913
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 30552968016
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)