Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC015170A_C01 KMC015170A_c01
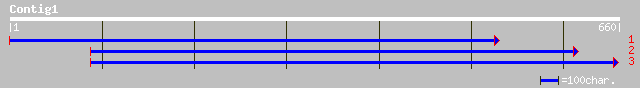
(660 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAK14411.1|AC087851_3 unknown protein [Oryza sativa] 181 1e-44
ref|NP_201245.1| putative protein; protein id: At5g64400.1, supp... 172 5e-42
ref|NP_196519.1| putative protein; protein id: At5g09570.1 [Arab... 151 7e-36
ref|NP_593716.1| conserved hypothetical protein [Schizosaccharom... 54 1e-06
gb|EAA35277.1| hypothetical protein [Neurospora crassa] 54 2e-06
>gb|AAK14411.1|AC087851_3 unknown protein [Oryza sativa]
Length = 138
Score = 181 bits (458), Expect = 1e-44
Identities = 87/131 (66%), Positives = 106/131 (80%), Gaps = 3/131 (2%)
Frame = -2
Query: 659 ARPAPRAAPRL---APVNHAPPPAPVQSSGGSMLGGLGATMAQGMALGTGSAIAHRAMDA 489
+RPAPRAAP P APPPAP Q+ GGS+LGG+G+T+AQGMA GTGSA+AHRA+DA
Sbjct: 9 SRPAPRAAPVRNPPQPARQAPPPAPAQNGGGSILGGIGSTIAQGMAFGTGSAMAHRAVDA 68
Query: 488 VMGPRVVQHETVAAEAAAAAPAPTAHAFGGDACGIHTKAFQDCINSYGADISKCQFYMDM 309
VMGPR +QHETV +EAAA+A P A+A D+C IH+KAFQDCIN+YG+DISKCQFY+DM
Sbjct: 69 VMGPRTIQHETVVSEAAASA-TPMANATPSDSCSIHSKAFQDCINNYGSDISKCQFYLDM 127
Query: 308 LAECRKNSGSS 276
L ECR+ S+
Sbjct: 128 LNECRRGGASA 138
>ref|NP_201245.1| putative protein; protein id: At5g64400.1, supported by cDNA:
gi_14334413, supported by cDNA: gi_17104660 [Arabidopsis
thaliana] gi|14334414|gb|AAK59405.1| unknown protein
[Arabidopsis thaliana] gi|17104661|gb|AAL34219.1|
unknown protein [Arabidopsis thaliana]
Length = 144
Score = 172 bits (435), Expect = 5e-42
Identities = 84/132 (63%), Positives = 100/132 (75%), Gaps = 2/132 (1%)
Frame = -2
Query: 656 RPAPRAAPRLAPVNHAPPPAPVQSSGGS--MLGGLGATMAQGMALGTGSAIAHRAMDAVM 483
RPA +P PV+ APPPAP Q+SGG M G+G+T+AQGMA GTGSA+AHRA+D+VM
Sbjct: 15 RPAAARSPAPQPVHRAPPPAPAQASGGGGGMFSGIGSTIAQGMAFGTGSAVAHRAVDSVM 74
Query: 482 GPRVVQHETVAAEAAAAAPAPTAHAFGGDACGIHTKAFQDCINSYGADISKCQFYMDMLA 303
GPR +QHE V EAA+A+ AP A C IH KAFQDCI SYG++ISKCQFYMDML+
Sbjct: 75 GPRTIQHEAV--EAASASAAPAGSAMLSSTCDIHAKAFQDCIGSYGSEISKCQFYMDMLS 132
Query: 302 ECRKNSGSSLSA 267
ECRKNSGS + A
Sbjct: 133 ECRKNSGSVIGA 144
>ref|NP_196519.1| putative protein; protein id: At5g09570.1 [Arabidopsis thaliana]
gi|11357507|pir||T49941 hypothetical protein F17I14.240
- Arabidopsis thaliana gi|7671432|emb|CAB89373.1|
putative protein [Arabidopsis thaliana]
gi|26452406|dbj|BAC43288.1| unknown protein [Arabidopsis
thaliana] gi|28372814|gb|AAO39889.1| At5g09570
[Arabidopsis thaliana]
Length = 139
Score = 151 bits (382), Expect = 7e-36
Identities = 74/132 (56%), Positives = 96/132 (72%), Gaps = 1/132 (0%)
Frame = -2
Query: 659 ARPAPRAAPRLAPVNHAPPPAPVQ-SSGGSMLGGLGATMAQGMALGTGSAIAHRAMDAVM 483
+RPA +P VN APPPA Q SSGGS LG +GA++ +G+A GTG+A HR +D+VM
Sbjct: 15 SRPAAARSPPPQSVNRAPPPATAQPSSGGSFLGNIGASITEGLAWGTGTAFGHRVVDSVM 74
Query: 482 GPRVVQHETVAAEAAAAAPAPTAHAFGGDACGIHTKAFQDCINSYGADISKCQFYMDMLA 303
GPR +HETV ++ +AA TA C IH+KAFQDC+N +G+DISKCQFYMDML+
Sbjct: 75 GPRTFKHETVVSQVPSAANTMTA-------CDIHSKAFQDCVNHFGSDISKCQFYMDMLS 127
Query: 302 ECRKNSGSSLSA 267
EC+KNSGS ++A
Sbjct: 128 ECKKNSGSVVAA 139
>ref|NP_593716.1| conserved hypothetical protein [Schizosaccharomyces pombe]
gi|1723468|sp|Q10307|YD52_SCHPO Hypothetical protein
C6C3.02c in chromosome I gi|7490297|pir||T39026
conserved hypothetical protein SPAC6C3.02c - fission
yeast (Schizosaccharomyces pombe)
gi|1204242|emb|CAA93615.1| hypothetical protein; similar
to S. cerevisiae YMR002W [Schizosaccharomyces pombe]
Length = 172
Score = 54.3 bits (129), Expect = 1e-06
Identities = 44/145 (30%), Positives = 60/145 (41%), Gaps = 23/145 (15%)
Frame = -2
Query: 659 ARPAPRAAPRLAP--VNHAPPPAPVQSSGGSMLGGLGATMAQGMALGTGSAIAHRAMDAV 486
A P AP AP V APPP VQ GGS G G ++ +G GSAI H +
Sbjct: 25 ALPPRTMAPPPAPSRVQQAPPPTAVQ--GGSSPGFFGNLVSTAAGVGIGSAIGHTVGSVI 82
Query: 485 MG----------------PRVVQHETVAAEAAAAAPAPTAHAF-----GGDACGIHTKAF 369
G P+ +V A +AP T + +AC K F
Sbjct: 83 TGGFSGSGSNNAPADTSVPQSSYSNSVPEAAYGSAPPSTFASSAISEEAKNACKGDAKMF 142
Query: 368 QDCINSYGADISKCQFYMDMLAECR 294
DCIN + + S+C +Y++ L C+
Sbjct: 143 ADCINEH--EFSQCSYYLEQLKACQ 165
>gb|EAA35277.1| hypothetical protein [Neurospora crassa]
Length = 450
Score = 53.9 bits (128), Expect = 2e-06
Identities = 38/121 (31%), Positives = 55/121 (45%)
Frame = -2
Query: 656 RPAPRAAPRLAPVNHAPPPAPVQSSGGSMLGGLGATMAQGMALGTGSAIAHRAMDAVMGP 477
RPA AP A HAPP A S G L G A+ A G+A+ GS+I H G
Sbjct: 327 RPASTYAPPAA-APHAPPAAAAPVSQGPGLFGQMASTAAGVAI--GSSIGHAIGGMFSGG 383
Query: 476 RVVQHETVAAEAAAAAPAPTAHAFGGDACGIHTKAFQDCINSYGADISKCQFYMDMLAEC 297
AA A A ++ G+ C TK+F C++ + ++ C +Y++ L C
Sbjct: 384 GSSAAPEAAAAPVQAQAAAAQNSSWGNNCSEATKSFTQCMDQHQGNMQICGWYLEQLKAC 443
Query: 296 R 294
+
Sbjct: 444 Q 444
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 588,286,445
Number of Sequences: 1393205
Number of extensions: 13703980
Number of successful extensions: 95812
Number of sequences better than 10.0: 215
Number of HSP's better than 10.0 without gapping: 60270
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 87756
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 28289785200
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)