Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC015100A_C01 KMC015100A_c01
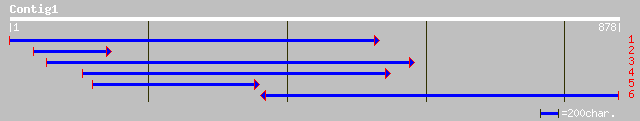
(877 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAD29731.1| protease inhibitor [Sesbania rostrata] 273 3e-72
prf||1802409A albumin 82 1e-14
sp|P32765|ASP_THECC 21 kDa seed protein precursor gi|99954|pir||... 82 1e-14
ref|NP_173228.1| lemir (miraculin), putative; protein id: At1g17... 79 7e-14
pir||T03803 tumor-related protein, clone NF34 - common tobacco g... 78 2e-13
>emb|CAD29731.1| protease inhibitor [Sesbania rostrata]
Length = 215
Score = 273 bits (697), Expect = 3e-72
Identities = 146/218 (66%), Positives = 167/218 (75%)
Frame = -3
Query: 836 QILPLCFLSLLVLNTQPLLADEPEPAVVDMQGNPLLPGVQYYVRPVSAEKGGLALGHTRN 657
Q LPLCFLSLLV NT PLLA EPEP VVD QG PL PGV YYV P+ A+ GGL LG TRN
Sbjct: 7 QFLPLCFLSLLVFNTHPLLAAEPEP-VVDKQGEPLQPGVGYYVWPLWADMGGLTLGQTRN 65
Query: 656 KTCPLDIILDPSSIIGSPTVFHTSSSSDVGYIPISTDLTVEMPILGSPCKEPKVWRLAKV 477
KTCPLD+I DPS IGSP FH + S D+G+IP TDLT+ +PILGS CKEPKVWRL+K
Sbjct: 66 KTCPLDVIRDPS-FIGSPVTFHVAGS-DLGFIPTLTDLTINIPILGSHCKEPKVWRLSKE 123
Query: 476 GSDFWFVSTQGVPGNLVSKFKIVRFEERDNHANEGEIYSFSFCPSVYGVICAPVGIYVDD 297
GS FWFVST GV G+L+SKFKI R E HA EIYSF FCPSV GV+CAPVG + D
Sbjct: 124 GSGFWFVSTGGVAGDLISKFKIERLE--GEHAY--EIYSFKFCPSVPGVLCAPVGTFEDA 179
Query: 296 DGTQVMAVGAGVGKPYHVRFQKVSSAYLELKNQDFSGV 183
DGT+VMAVG G+ +PY+VRFQK S++Y + K Q+FS V
Sbjct: 180 DGTKVMAVGDGI-EPYYVRFQK-STSYSQKKGQEFSSV 215
>prf||1802409A albumin
Length = 221
Score = 81.6 bits (200), Expect = 1e-14
Identities = 69/204 (33%), Positives = 95/204 (45%), Gaps = 15/204 (7%)
Frame = -3
Query: 857 MKLGTMFQILPLCFLSLLVLNTQPLLADEPEPAVVDMQGNPLLPGVQYYVRPV--SAEKG 684
MK T +L F S A+ P V+D G+ L GVQYYV A G
Sbjct: 1 MKTATAVVLLLFAFTSKSYFFGVANAANSP---VLDTDGDELQTGVQYYVLSSISGAGGG 57
Query: 683 GLALGHTRNKTCPLDIILDPSSI-IGSPTVFHTSSSSDVGYIPISTDLTVE-MPILGSPC 510
GLALG ++CP ++ S + G+P +F + S D + +STD+ +E +PI C
Sbjct: 58 GLALGRATGQSCPEIVVQRRSDLDNGTPVIFSNADSKD-DVVRVSTDVNIEFVPIRDRLC 116
Query: 509 KEPKVWRLAKVGSDF--WFVSTQGV-----PGNLVSKFKIVRFEERDNHANEGEI-YSFS 354
VWRL + W+V+T GV P L S FKI + G + Y F
Sbjct: 117 STSTVWRLDNYDNSAGKWWVTTDGVKGEPGPNTLCSWFKIEK---------AGVLGYKFR 167
Query: 353 FCPSVYG---VICAPVGIYVDDDG 291
FCPSV +C+ +G + DDDG
Sbjct: 168 FCPSVCDSCTTLCSDIGRHSDDDG 191
>sp|P32765|ASP_THECC 21 kDa seed protein precursor gi|99954|pir||S16252 trypsin
inhibitor homolog - soybean gi|21909|emb|CAA39860.1| 21
kDa seed protein [Theobroma cacao]
Length = 221
Score = 81.6 bits (200), Expect = 1e-14
Identities = 69/204 (33%), Positives = 95/204 (45%), Gaps = 15/204 (7%)
Frame = -3
Query: 857 MKLGTMFQILPLCFLSLLVLNTQPLLADEPEPAVVDMQGNPLLPGVQYYVRPV--SAEKG 684
MK T +L F S A+ P V+D G+ L GVQYYV A G
Sbjct: 1 MKTATAVVLLLFAFTSKSYFFGVANAANSP---VLDTDGDELQTGVQYYVLSSISGAGGG 57
Query: 683 GLALGHTRNKTCPLDIILDPSSI-IGSPTVFHTSSSSDVGYIPISTDLTVE-MPILGSPC 510
GLALG ++CP ++ S + G+P +F + S D + +STD+ +E +PI C
Sbjct: 58 GLALGRATGQSCPEIVVQRRSDLDNGTPVIFSNADSKD-DVVRVSTDVNIEFVPIRDRLC 116
Query: 509 KEPKVWRLAKVGSDF--WFVSTQGV-----PGNLVSKFKIVRFEERDNHANEGEI-YSFS 354
VWRL + W+V+T GV P L S FKI + G + Y F
Sbjct: 117 STSTVWRLDNYDNSAGKWWVTTDGVKGEPGPNTLCSWFKIEK---------AGVLGYKFR 167
Query: 353 FCPSVYG---VICAPVGIYVDDDG 291
FCPSV +C+ +G + DDDG
Sbjct: 168 FCPSVCDSCTTLCSDIGRHSDDDG 191
>ref|NP_173228.1| lemir (miraculin), putative; protein id: At1g17860.1, supported by
cDNA: gi_12083239, supported by cDNA: gi_13899080,
supported by cDNA: gi_15294165, supported by cDNA:
gi_20148400, supported by cDNA: gi_20453292 [Arabidopsis
thaliana] gi|25294073|pir||G86313 hypothetical protein
F2H15.9 - Arabidopsis thaliana
gi|9665064|gb|AAF97266.1|AC034106_9 Contains similarity
to a tumor-related protein from Nicotiana tabacum
gb|U66263 and contains a trypsin and protease inhibitor
PF|00197 domain. ESTs gb|AV561824, gb|T44961,
gb|H36186, gb|T45060, gb|N38006, gb|F19847 come from
this gene. [Arabidopsis thaliana]
gi|12083240|gb|AAG48779.1|AF332416_1 putative lemir
(miraculin) protein [Arabidopsis thaliana]
gi|13899081|gb|AAK48962.1|AF370535_1 Unknown protein
[Arabidopsis thaliana]
gi|15294166|gb|AAK95260.1|AF410274_1 At1g17860/F2H15_8
[Arabidopsis thaliana] gi|20148401|gb|AAM10091.1|
unknown protein [Arabidopsis thaliana]
gi|20453293|gb|AAM19885.1| At1g17860/F2H15_8
[Arabidopsis thaliana]
Length = 196
Score = 79.3 bits (194), Expect = 7e-14
Identities = 62/194 (31%), Positives = 90/194 (45%), Gaps = 12/194 (6%)
Frame = -3
Query: 842 MFQILPLCFLSLLVLNTQPLLADEPEPAVVDMQGNPLLPGVQYYVRPV-SAEKGGLALGH 666
M +L + L + ++ + + + V D+ G LL GV YY+ PV GGL + +
Sbjct: 1 MSSLLYIFLLLAVFISHRGVTTEAAVEPVKDINGKSLLTGVNYYILPVIRGRGGGLTMSN 60
Query: 665 TRNKTCPLDIILDPSSII-GSPTVFHTSSSSDVGYIPISTDLTVEMPILGSPCKEPKVWR 489
+ +TCP +I D + G P F S IP+STD+ ++ SP +W
Sbjct: 61 LKTETCPTSVIQDQFEVSQGLPVKFSPYDKSRT--IPVSTDVNIKF----SP---TSIWE 111
Query: 488 LAKVG--SDFWFVSTQGVPGNLVSK-----FKIVRFEERDNHANEGEIYSFSFCPSVYG- 333
LA + WF+ST GV GN K FKI +FE+ Y FCP+V
Sbjct: 112 LANFDETTKQWFISTCGVEGNPGQKTVDNWFKIDKFEKD---------YKIRFCPTVCNF 162
Query: 332 --VICAPVGIYVDD 297
VIC VG++V D
Sbjct: 163 CKVICRDVGVFVQD 176
>pir||T03803 tumor-related protein, clone NF34 - common tobacco
gi|1762933|gb|AAC49969.1| tumor-related protein
[Nicotiana tabacum]
Length = 210
Score = 78.2 bits (191), Expect = 2e-13
Identities = 68/198 (34%), Positives = 96/198 (48%), Gaps = 17/198 (8%)
Frame = -3
Query: 773 EPEPAVVDMQGNPLLPGVQYYVRP-VSAEKGGLALGHTRNKTCPLDIILDPSSII--GSP 603
E PAVVD+ G L G+ YY+ P V GGL L T N++CPLD ++ I G P
Sbjct: 26 EAPPAVVDIAGKKLRTGIDYYILPVVRGRGGGLTLDSTGNESCPLDAVVQEQQEIKNGLP 85
Query: 602 TVFHTSSSSDVGYIPISTDLTVEMPILGSPCKEPKVWRLAKVGSDF------WFVSTQGV 441
F T + G I STDL ++ S C + +W+L DF +F++ G
Sbjct: 86 LTF-TPVNPKKGVIRESTDLNIKFS-AASICVQTTLWKL----DDFDETTGKYFITIGGN 139
Query: 440 PGN-----LVSKFKIVRFEERDNHANEGEIYSFSFCPSVYG---VICAPVGIYVDDDGTQ 285
GN + + FKI +F ERD Y +CP+V VIC VGI++ DG +
Sbjct: 140 EGNPGRETISNWFKIEKF-ERD--------YKLVYCPTVCNFCKVICKDVGIFI-QDGIR 189
Query: 284 VMAVGAGVGKPYHVRFQK 231
+A+ P+ V F+K
Sbjct: 190 RLALS---DVPFKVMFKK 204
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 784,231,402
Number of Sequences: 1393205
Number of extensions: 18444063
Number of successful extensions: 53784
Number of sequences better than 10.0: 183
Number of HSP's better than 10.0 without gapping: 48078
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 52667
length of database: 448,689,247
effective HSP length: 122
effective length of database: 278,718,237
effective search space used: 47103382053
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)