Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
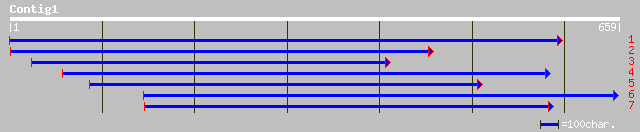
Query= KMC015048A_C01 KMC015048A_c01
(657 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_198495.1| p-nitrophenylphosphatase-like protein; protein ... 228 5e-59
ref|NP_198485.1| N-glyceraldehyde-2-phosphotransferase-like; pro... 228 5e-59
dbj|BAC42546.1| putative p-nitrophenylphosphatase [Arabidopsis t... 228 5e-59
emb|CAD41136.1| OSJNBa0084K20.16 [Oryza sativa (japonica cultiva... 221 5e-57
dbj|BAA98057.1| 4-nitrophenylphosphatase-like [Arabidopsis thali... 180 8e-47
>ref|NP_198495.1| p-nitrophenylphosphatase-like protein; protein id: At5g36790.1,
supported by cDNA: gi_20453152 [Arabidopsis thaliana]
gi|20453153|gb|AAM19818.1| AT5g36790/f5h8_20
[Arabidopsis thaliana] gi|21689621|gb|AAM67432.1|
AT5g36790/f5h8_20 [Arabidopsis thaliana]
Length = 362
Score = 228 bits (581), Expect = 5e-59
Identities = 113/123 (91%), Positives = 117/123 (94%)
Frame = -1
Query: 657 LFIATNRDAVTHLTDAQEWAGGGSMVGAFVGSTQREPLVVGKPSTFMMDYLANEFGISKS 478
LFIATNRDAVTHLTDAQEWAGGGSMVGA VGSTQREPLVVGKPSTFMMDYLA++FGI KS
Sbjct: 240 LFIATNRDAVTHLTDAQEWAGGGSMVGALVGSTQREPLVVGKPSTFMMDYLADKFGIQKS 299
Query: 477 QICMVGDRLDTDILFGQNGGCKTLLVLSGVTSLSMLQSPNNSIQPDFYTSKISDFLSLKA 298
QICMVGDRLDTDILFGQNGGCKTLLVLSGVTS+SML+SP N IQPDFYTSKISDFLS KA
Sbjct: 300 QICMVGDRLDTDILFGQNGGCKTLLVLSGVTSISMLESPENKIQPDFYTSKISDFLSPKA 359
Query: 297 AAV 289
A V
Sbjct: 360 ATV 362
>ref|NP_198485.1| N-glyceraldehyde-2-phosphotransferase-like; protein id: At5g36700.1
[Arabidopsis thaliana] gi|8885622|dbj|BAA97552.1|
N-glyceraldehyde-2-phosphotransferase-like [Arabidopsis
thaliana]
Length = 289
Score = 228 bits (581), Expect = 5e-59
Identities = 113/123 (91%), Positives = 117/123 (94%)
Frame = -1
Query: 657 LFIATNRDAVTHLTDAQEWAGGGSMVGAFVGSTQREPLVVGKPSTFMMDYLANEFGISKS 478
LFIATNRDAVTHLTDAQEWAGGGSMVGA VGSTQREPLVVGKPSTFMMDYLA++FGI KS
Sbjct: 167 LFIATNRDAVTHLTDAQEWAGGGSMVGALVGSTQREPLVVGKPSTFMMDYLADKFGIQKS 226
Query: 477 QICMVGDRLDTDILFGQNGGCKTLLVLSGVTSLSMLQSPNNSIQPDFYTSKISDFLSLKA 298
QICMVGDRLDTDILFGQNGGCKTLLVLSGVTS+SML+SP N IQPDFYTSKISDFLS KA
Sbjct: 227 QICMVGDRLDTDILFGQNGGCKTLLVLSGVTSISMLESPENKIQPDFYTSKISDFLSPKA 286
Query: 297 AAV 289
A V
Sbjct: 287 ATV 289
>dbj|BAC42546.1| putative p-nitrophenylphosphatase [Arabidopsis thaliana]
Length = 309
Score = 228 bits (581), Expect = 5e-59
Identities = 113/123 (91%), Positives = 117/123 (94%)
Frame = -1
Query: 657 LFIATNRDAVTHLTDAQEWAGGGSMVGAFVGSTQREPLVVGKPSTFMMDYLANEFGISKS 478
LFIATNRDAVTHLTDAQEWAGGGSMVGA VGSTQREPLVVGKPSTFMMDYLA++FGI KS
Sbjct: 187 LFIATNRDAVTHLTDAQEWAGGGSMVGALVGSTQREPLVVGKPSTFMMDYLADKFGIQKS 246
Query: 477 QICMVGDRLDTDILFGQNGGCKTLLVLSGVTSLSMLQSPNNSIQPDFYTSKISDFLSLKA 298
QICMVGDRLDTDILFGQNGGCKTLLVLSGVTS+SML+SP N IQPDFYTSKISDFLS KA
Sbjct: 247 QICMVGDRLDTDILFGQNGGCKTLLVLSGVTSISMLESPENKIQPDFYTSKISDFLSPKA 306
Query: 297 AAV 289
A V
Sbjct: 307 ATV 309
>emb|CAD41136.1| OSJNBa0084K20.16 [Oryza sativa (japonica cultivar-group)]
Length = 372
Score = 221 bits (564), Expect = 5e-57
Identities = 108/123 (87%), Positives = 118/123 (95%)
Frame = -1
Query: 657 LFIATNRDAVTHLTDAQEWAGGGSMVGAFVGSTQREPLVVGKPSTFMMDYLANEFGISKS 478
LFIATNRDAVTHLTDAQEWAGGGSMVGA +GST++EPLVVGKPSTFMMDYLA +FGI+ S
Sbjct: 250 LFIATNRDAVTHLTDAQEWAGGGSMVGAILGSTKQEPLVVGKPSTFMMDYLAKKFGITTS 309
Query: 477 QICMVGDRLDTDILFGQNGGCKTLLVLSGVTSLSMLQSPNNSIQPDFYTSKISDFLSLKA 298
QICMVGDRLDTDILFGQNGGCKTLLVLSGVTS+ MLQSP+NSIQPDFYT++ISDFL+LKA
Sbjct: 310 QICMVGDRLDTDILFGQNGGCKTLLVLSGVTSVQMLQSPDNSIQPDFYTNQISDFLTLKA 369
Query: 297 AAV 289
A V
Sbjct: 370 ATV 372
>dbj|BAA98057.1| 4-nitrophenylphosphatase-like [Arabidopsis thaliana]
Length = 389
Score = 180 bits (456), Expect(2) = 8e-47
Identities = 87/93 (93%), Positives = 91/93 (97%)
Frame = -1
Query: 657 LFIATNRDAVTHLTDAQEWAGGGSMVGAFVGSTQREPLVVGKPSTFMMDYLANEFGISKS 478
LFIATNRDAVTHLTDAQEWAGGGSMVGA VGSTQREPLVVGKPSTFMMDYLA++FGI KS
Sbjct: 240 LFIATNRDAVTHLTDAQEWAGGGSMVGALVGSTQREPLVVGKPSTFMMDYLADKFGIQKS 299
Query: 477 QICMVGDRLDTDILFGQNGGCKTLLVLSGVTSL 379
QICMVGDRLDTDILFGQNGGCKTLLVLSG+T+L
Sbjct: 300 QICMVGDRLDTDILFGQNGGCKTLLVLSGITNL 332
Score = 29.3 bits (64), Expect(2) = 8e-47
Identities = 15/30 (50%), Positives = 20/30 (66%)
Frame = -2
Query: 374 CFRALTTPSSQISTQAKFQIFFPSKLQLYD 285
C++AL T +QIST A+ IF K QLY+
Sbjct: 351 CWKALRTRYNQISTPARSPIFCLRKPQLYN 380
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 574,430,827
Number of Sequences: 1393205
Number of extensions: 12443223
Number of successful extensions: 27973
Number of sequences better than 10.0: 211
Number of HSP's better than 10.0 without gapping: 26815
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 27921
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 28006887348
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)