Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC014968A_C01 KMC014968A_c01
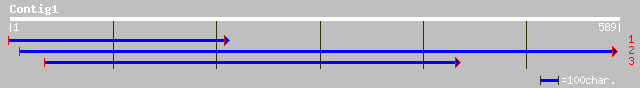
(589 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM18188.2| deetiolated 1-like protein [Solanum tuberosum] 249 1e-65
emb|CAA10993.1| tDET1 protein [Lycopersicon esculentum] gi|44543... 246 1e-64
ref|NP_192756.1| Deetiolated1 (DET1) light signal transduction p... 230 1e-59
dbj|BAB16336.1| putative tDET1 protein [Oryza sativa (japonica c... 212 2e-54
gb|AAO50779.1| similar to Mus musculus (Mouse). Sex-determining ... 92 5e-18
>gb|AAM18188.2| deetiolated 1-like protein [Solanum tuberosum]
Length = 524
Score = 249 bits (637), Expect = 1e-65
Identities = 123/146 (84%), Positives = 134/146 (91%)
Frame = -3
Query: 587 SRNSMYMNFISSHSNNIHALEQLRSIKDKASNSSQFVKKMLASLPFSCQSQSPSPYFDQS 408
S++S++MNF+SSHSNNIHALEQLR K+KA+N SQFVKKM+ASLPFSCQSQSPSPYFDQS
Sbjct: 379 SKSSLHMNFMSSHSNNIHALEQLRCTKNKATNFSQFVKKMMASLPFSCQSQSPSPYFDQS 438
Query: 407 LFRFDDKLISATDRHRQSTDHPIKFILRKPPYSLKFKIKPGPEAGSMDGRAKKISSFLFH 228
LFRFD+KLISA DRHRQSTDHPIKFI R+ P LKFK+KPGPEAGS DGR KKI SFLFH
Sbjct: 439 LFRFDEKLISAIDRHRQSTDHPIKFISRRQPNILKFKMKPGPEAGSTDGRTKKICSFLFH 498
Query: 227 PILPLALSVQQTLFLQPSVVNIHFRR 150
PILPLALSVQQTLFLQ SVVNIHFRR
Sbjct: 499 PILPLALSVQQTLFLQASVVNIHFRR 524
>emb|CAA10993.1| tDET1 protein [Lycopersicon esculentum] gi|4454332|emb|CAA11914.1|
tDET1 protein [Lycopersicon esculentum]
Length = 523
Score = 246 bits (629), Expect = 1e-64
Identities = 122/146 (83%), Positives = 133/146 (90%)
Frame = -3
Query: 587 SRNSMYMNFISSHSNNIHALEQLRSIKDKASNSSQFVKKMLASLPFSCQSQSPSPYFDQS 408
S++S++MNF+SSHSNNIHALEQLR K+KA+N SQFVKKM+ASLP SCQSQSPSPYFDQS
Sbjct: 378 SKSSLHMNFMSSHSNNIHALEQLRCTKNKATNFSQFVKKMMASLPCSCQSQSPSPYFDQS 437
Query: 407 LFRFDDKLISATDRHRQSTDHPIKFILRKPPYSLKFKIKPGPEAGSMDGRAKKISSFLFH 228
LFRFD+KLISA DRHRQSTDHPIKFI R+ P LKFK+KPGPEAGS DGR KKI SFLFH
Sbjct: 438 LFRFDEKLISAIDRHRQSTDHPIKFISRRQPNILKFKMKPGPEAGSTDGRTKKICSFLFH 497
Query: 227 PILPLALSVQQTLFLQPSVVNIHFRR 150
PILPLALSVQQTLFLQ SVVNIHFRR
Sbjct: 498 PILPLALSVQQTLFLQASVVNIHFRR 523
>ref|NP_192756.1| Deetiolated1 (DET1) light signal transduction protein; protein id:
At4g10180.1 [Arabidopsis thaliana]
gi|1352243|sp|P48732|DET1_ARATH Light-mediated
development protein DET1 (Deetiolated1)
gi|625972|pir||A54841 DET1 protein - Arabidopsis
thaliana gi|510275|gb|AAB59299.1| DET1
gi|3695414|gb|AAC62814.1| Arabidopsis thaliana
light-mediated development protein DET1 (SP:P48732)
gi|4538982|emb|CAB39770.1| Deetiolated1 (DET1) light
signal transduction protein [Arabidopsis thaliana]
gi|7267714|emb|CAB78141.1| Deetiolated1 (DET1) light
signal transduction protein [Arabidopsis thaliana]
Length = 541
Score = 230 bits (586), Expect = 1e-59
Identities = 109/146 (74%), Positives = 131/146 (89%)
Frame = -3
Query: 587 SRNSMYMNFISSHSNNIHALEQLRSIKDKASNSSQFVKKMLASLPFSCQSQSPSPYFDQS 408
S ++ +MNF++SHSNN++ALEQL+ K+K+++ SQFVKKML SLPFSCQSQSPSPYFDQS
Sbjct: 396 SSSTPFMNFVTSHSNNVYALEQLKYTKNKSNSFSQFVKKMLLSLPFSCQSQSPSPYFDQS 455
Query: 407 LFRFDDKLISATDRHRQSTDHPIKFILRKPPYSLKFKIKPGPEAGSMDGRAKKISSFLFH 228
LFRFD+KLISA DRHRQS+D+PIKFI R+ P +LKFKIKPGPE G+ DGR+KKI SFLFH
Sbjct: 456 LFRFDEKLISAADRHRQSSDNPIKFISRRQPQTLKFKIKPGPECGTADGRSKKICSFLFH 515
Query: 227 PILPLALSVQQTLFLQPSVVNIHFRR 150
P LPLA+S+QQTLF+ PSVVNIHFRR
Sbjct: 516 PHLPLAISIQQTLFMPPSVVNIHFRR 541
>dbj|BAB16336.1| putative tDET1 protein [Oryza sativa (japonica cultivar-group)]
Length = 512
Score = 212 bits (540), Expect = 2e-54
Identities = 104/146 (71%), Positives = 126/146 (86%), Gaps = 1/146 (0%)
Frame = -3
Query: 584 RNSMYMNFISSHSNNIHALEQLRSIKDKA-SNSSQFVKKMLASLPFSCQSQSPSPYFDQS 408
+NS + NFISSHSNN+HAL+QLR+IK+KA S S QFVKKM+ASLP++CQSQSPSPYFD S
Sbjct: 368 QNSSHGNFISSHSNNVHALDQLRTIKNKANSTSQQFVKKMMASLPYTCQSQSPSPYFDLS 427
Query: 407 LFRFDDKLISATDRHRQSTDHPIKFILRKPPYSLKFKIKPGPEAGSMDGRAKKISSFLFH 228
LFR+D+KLISA DRHR T+HPIKFI K P +KFKIKPG ++G+ D RAK+ISSFLFH
Sbjct: 428 LFRYDEKLISAIDRHRHCTEHPIKFISVKQPNVVKFKIKPGSDSGASDSRAKRISSFLFH 487
Query: 227 PILPLALSVQQTLFLQPSVVNIHFRR 150
P PLALS+QQT ++QP+VVN+HFRR
Sbjct: 488 PFFPLALSIQQT-YMQPTVVNLHFRR 512
>gb|AAO50779.1| similar to Mus musculus (Mouse). Sex-determining region Y protein
(Testis-determining factor) [Dictyostelium discoideum]
Length = 1461
Score = 92.0 bits (227), Expect = 5e-18
Identities = 52/143 (36%), Positives = 85/143 (59%), Gaps = 5/143 (3%)
Frame = -3
Query: 569 MNFISSHSNNI----HALEQ-LRSIKDKASNSSQFVKKMLASLPFSCQSQSPSPYFDQSL 405
+ F S+ SNN H ++Q L + + +Q VK++L++LPFS QS + SPYFD+ L
Sbjct: 1280 IKFRSTCSNNSFVREHLIKQKLAVVNARYGGETQAVKRILSALPFSPQSWNESPYFDKYL 1339
Query: 404 FRFDDKLISATDRHRQSTDHPIKFILRKPPYSLKFKIKPGPEAGSMDGRAKKISSFLFHP 225
F +D+K+IS+T+R + D+PIKF R +KFK+ G ++ + KK ++++FHP
Sbjct: 1340 FSYDEKIISSTERPKPLVDYPIKFYSRVSG-EVKFKLNTGGAQEKIERQNKKYATYIFHP 1398
Query: 224 ILPLALSVQQTLFLQPSVVNIHF 156
P +S+ + + VN HF
Sbjct: 1399 YYPFIISLLHSPSTGSAAVNFHF 1421
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 452,469,551
Number of Sequences: 1393205
Number of extensions: 9089750
Number of successful extensions: 24814
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 23943
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24802
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 22283372436
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)