Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC014903A_C01 KMC014903A_c01
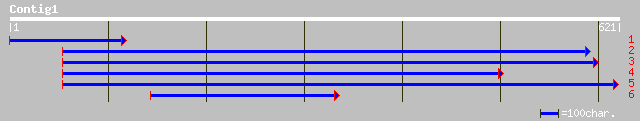
(621 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM13903.1| putative cytochrome P450 [Arabidopsis thaliana] 191 5e-48
ref|NP_190881.1| cytochrome p450 family; protein id: At3g53130.1... 191 5e-48
gb|AAK20054.1|AC025783_14 putative cytochrome P450 monooxygenase... 179 3e-44
pir||F86441 probable cytochrome P450 [imported] - Arabidopsis th... 127 2e-28
ref|NP_564384.1| cytochrome P450, putative; protein id: At1g3180... 127 2e-28
>gb|AAM13903.1| putative cytochrome P450 [Arabidopsis thaliana]
Length = 552
Score = 191 bits (486), Expect = 5e-48
Identities = 89/105 (84%), Positives = 99/105 (93%)
Frame = -2
Query: 620 DIMISVYNIHHSSEVWDRAEEFLPERFDLDGPMPNETNTDFRFIPFSGGPRKCVGDQFAL 441
DIMISVYNIH SSEVW++AEEFLPERFD+DG +PNETNTDF+FIPFSGGPRKCVGDQFAL
Sbjct: 448 DIMISVYNIHRSSEVWEKAEEFLPERFDIDGAIPNETNTDFKFIPFSGGPRKCVGDQFAL 507
Query: 440 LEATVSLAIFLQHMNFELVPDQNISMTTGATIHTTNGLYMKLSQR 306
+EA V+LA+FLQ +N ELVPDQ ISMTTGATIHTTNGLYMK+SQR
Sbjct: 508 MEAIVALAVFLQRLNVELVPDQTISMTTGATIHTTNGLYMKVSQR 552
>ref|NP_190881.1| cytochrome p450 family; protein id: At3g53130.1 [Arabidopsis
thaliana] gi|11249712|pir||T46159 cytochrome P450-like
protein - Arabidopsis thaliana
gi|6630733|emb|CAB64216.1| Cytochrom P450-like protein
[Arabidopsis thaliana]
Length = 566
Score = 191 bits (486), Expect = 5e-48
Identities = 89/105 (84%), Positives = 99/105 (93%)
Frame = -2
Query: 620 DIMISVYNIHHSSEVWDRAEEFLPERFDLDGPMPNETNTDFRFIPFSGGPRKCVGDQFAL 441
DIMISVYNIH SSEVW++AEEFLPERFD+DG +PNETNTDF+FIPFSGGPRKCVGDQFAL
Sbjct: 462 DIMISVYNIHRSSEVWEKAEEFLPERFDIDGAIPNETNTDFKFIPFSGGPRKCVGDQFAL 521
Query: 440 LEATVSLAIFLQHMNFELVPDQNISMTTGATIHTTNGLYMKLSQR 306
+EA V+LA+FLQ +N ELVPDQ ISMTTGATIHTTNGLYMK+SQR
Sbjct: 522 MEAIVALAVFLQRLNVELVPDQTISMTTGATIHTTNGLYMKVSQR 566
>gb|AAK20054.1|AC025783_14 putative cytochrome P450 monooxygenase [Oryza sativa (japonica
cultivar-group)]
Length = 584
Score = 179 bits (453), Expect = 3e-44
Identities = 82/102 (80%), Positives = 94/102 (91%)
Frame = -2
Query: 620 DIMISVYNIHHSSEVWDRAEEFLPERFDLDGPMPNETNTDFRFIPFSGGPRKCVGDQFAL 441
DIMISVYNIH S EVWDRA++F+PERFDL+GP+PNETNT++RFIPFSGGPRKCVGDQFAL
Sbjct: 443 DIMISVYNIHRSPEVWDRADDFIPERFDLEGPVPNETNTEYRFIPFSGGPRKCVGDQFAL 502
Query: 440 LEATVSLAIFLQHMNFELVPDQNISMTTGATIHTTNGLYMKL 315
LEA V+LA+ LQ M+ ELVPDQ I+MTTGATIHTTNGLYM +
Sbjct: 503 LEAIVALAVVLQKMDIELVPDQKINMTTGATIHTTNGLYMNV 544
>pir||F86441 probable cytochrome P450 [imported] - Arabidopsis thaliana
gi|12321297|gb|AAG50718.1|AC079041_11 cytochrome P450,
putative [Arabidopsis thaliana]
Length = 593
Score = 127 bits (318), Expect = 2e-28
Identities = 58/108 (53%), Positives = 78/108 (71%), Gaps = 1/108 (0%)
Frame = -2
Query: 620 DIMISVYNIHHSSEVWDRAEEFLPERFDLDGPMPNETNTDFRFIPFSGGPRKCVGDQFAL 441
DI ISV+N+H S WD AE+F PER+ LDGP PNETN +F ++PF GGPRKC+GD FA
Sbjct: 462 DIFISVWNLHRSPLHWDDAEKFNPERWPLDGPNPNETNQNFSYLPFGGGPRKCIGDMFAS 521
Query: 440 LEATVSLAIFLQHMNFELVPD-QNISMTTGATIHTTNGLYMKLSQRVK 300
E V++A+ ++ NF++ P + MTTGATIHTT GL + +++R K
Sbjct: 522 FENVVAIAMLIRRFNFQIAPGAPPVKMTTGATIHTTEGLKLTVTKRTK 569
>ref|NP_564384.1| cytochrome P450, putative; protein id: At1g31800.1, supported by
cDNA: gi_15912336, supported by cDNA: gi_16648792
[Arabidopsis thaliana] gi|15912337|gb|AAL08302.1|
At1g31800/68069_m00159 [Arabidopsis thaliana]
gi|16648793|gb|AAL25587.1| At1g31800/68069_m00159
[Arabidopsis thaliana] gi|22655378|gb|AAM98281.1|
At1g31800/68069_m00159 [Arabidopsis thaliana]
Length = 595
Score = 127 bits (318), Expect = 2e-28
Identities = 58/108 (53%), Positives = 78/108 (71%), Gaps = 1/108 (0%)
Frame = -2
Query: 620 DIMISVYNIHHSSEVWDRAEEFLPERFDLDGPMPNETNTDFRFIPFSGGPRKCVGDQFAL 441
DI ISV+N+H S WD AE+F PER+ LDGP PNETN +F ++PF GGPRKC+GD FA
Sbjct: 464 DIFISVWNLHRSPLHWDDAEKFNPERWPLDGPNPNETNQNFSYLPFGGGPRKCIGDMFAS 523
Query: 440 LEATVSLAIFLQHMNFELVPD-QNISMTTGATIHTTNGLYMKLSQRVK 300
E V++A+ ++ NF++ P + MTTGATIHTT GL + +++R K
Sbjct: 524 FENVVAIAMLIRRFNFQIAPGAPPVKMTTGATIHTTEGLKLTVTKRTK 571
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 500,966,802
Number of Sequences: 1393205
Number of extensions: 10166273
Number of successful extensions: 27118
Number of sequences better than 10.0: 2339
Number of HSP's better than 10.0 without gapping: 25169
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25981
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25017613016
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)