Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC014832A_C01 KMC014832A_c01
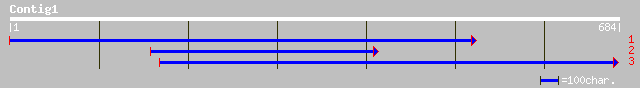
(684 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||S69190 myb-related protein 1 - tomato gi|1167486|emb|CAA646... 43 0.004
ref|NP_176575.1| putative MYB family transcription factor; prote... 37 0.25
ref|NP_179575.1| hypothetical protein; protein id: At2g19850.1 [... 33 3.7
>pir||S69190 myb-related protein 1 - tomato gi|1167486|emb|CAA64615.1|
transcription factor [Lycopersicon esculentum]
Length = 331
Score = 43.1 bits (100), Expect = 0.004
Identities = 29/58 (50%), Positives = 35/58 (60%), Gaps = 6/58 (10%)
Frame = -3
Query: 676 LNEW---VETQQQQCPNFLFWDNVEGHFGGGE--LAP-NSSNMGTITNTTLSPFPSSL 521
LNEW V+TQQ CP++LFWD G GG + P S+NMG I LS FPS+L
Sbjct: 279 LNEWNSQVDTQQC-CPSYLFWDQENGSIGGDHEIIDPTTSNNMGQI----LSSFPSTL 331
>ref|NP_176575.1| putative MYB family transcription factor; protein id: At1g63910.1,
supported by cDNA: gi_15375284 [Arabidopsis thaliana]
gi|25294100|pir||C96664 hypothetical protein T12P18.7
[imported] - Arabidopsis thaliana
gi|6708470|gb|AAF25949.1|AF214116_1 putative
transcription factor [Arabidopsis thaliana]
gi|12325018|gb|AAG52460.1|AC010852_17 putative MYB
family transcription factor; 19087-20744 [Arabidopsis
thaliana]
Length = 370
Score = 37.0 bits (84), Expect = 0.25
Identities = 22/56 (39%), Positives = 31/56 (55%), Gaps = 3/56 (5%)
Frame = -3
Query: 682 LHLNEWVETQQQQCPNFLFWDNVEGHFGGGELAPN---SSNMGTITNTTLSPFPSS 524
L ++W ++QQ C NF FWDN+ + G L N S N+G ++ S FPSS
Sbjct: 318 LSYDQWDDSQQ--CSNFFFWDNLNINVEGSSLVGNQDPSMNLG--SSALSSSFPSS 369
>ref|NP_179575.1| hypothetical protein; protein id: At2g19850.1 [Arabidopsis
thaliana] gi|25411936|pir||H84581 hypothetical protein
At2g19850 [imported] - Arabidopsis thaliana
gi|3687233|gb|AAC62131.1| hypothetical protein
[Arabidopsis thaliana]
Length = 296
Score = 33.1 bits (74), Expect = 3.7
Identities = 19/50 (38%), Positives = 25/50 (50%), Gaps = 5/50 (10%)
Frame = +3
Query: 546 VVFVIVPIF-DEF----GASSPPPKCPSTLSQKRKLGHCCCWVSTHSFKC 680
V + IV +F D F +SP + PS LS RKL CC + +F C
Sbjct: 205 VAYAIVAVFHDAFYPLEDVASPDSRSPSVLSYFRKLISCCSTIGVSNFSC 254
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 578,441,663
Number of Sequences: 1393205
Number of extensions: 12149640
Number of successful extensions: 29443
Number of sequences better than 10.0: 6
Number of HSP's better than 10.0 without gapping: 28304
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 29435
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 30552968016
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)