Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC014816A_C01 KMC014816A_c01
(611 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
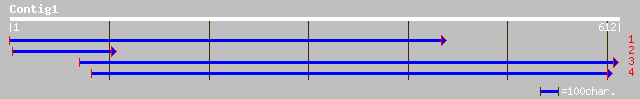
ref|NP_180609.1| unknown protein; protein id: At2g30510.1 [Arabi... 169 3e-41
gb|AAK59568.1| unknown protein [Arabidopsis thaliana] gi|1581064... 169 3e-41
gb|AAF33112.1|AF181683_1 RPT2 [Arabidopsis thaliana] 167 8e-41
gb|AAO16688.1| RPT2-like protein [Sorghum bicolor] 86 3e-16
gb|AAN77296.1| Hypothetical protein [Oryza sativa (japonica cult... 76 4e-13
>ref|NP_180609.1| unknown protein; protein id: At2g30510.1 [Arabidopsis thaliana]
gi|25408081|pir||C84709 hypothetical protein At2g30510
[imported] - Arabidopsis thaliana
gi|1946368|gb|AAB63086.1| unknown protein [Arabidopsis
thaliana]
Length = 415
Score = 169 bits (427), Expect = 3e-41
Identities = 99/156 (63%), Positives = 114/156 (72%), Gaps = 23/156 (14%)
Frame = -1
Query: 611 LKLSYEARVHASQNKRLPVQIVLHALYYDQLRLRSGADDG--------GEAVAEREQLQT 456
LKLSY+AR+HASQNKRLPV IVLHALYYDQL+LRSG + EA+ R QLQ
Sbjct: 260 LKLSYDARLHASQNKRLPVNIVLHALYYDQLKLRSGVAEQEERAVVVLPEALKTRSQLQA 319
Query: 455 DVTLVRENEELRLELMRMKTVVSDLQK-----GVAPS------SSKKIK--FFSSVSKKL 315
D TL +ENE LR ELM+MK VSD+QK G + S SSKK K FFSSVSKKL
Sbjct: 320 DTTLAKENEALRSELMKMKMYVSDMQKNKNGAGASSSNSSSLVSSKKSKHTFFSSVSKKL 379
Query: 314 GKLNPFRNGSKDTTHLED--GPVDLTKPRRRRFSIS 213
GKLNPF+NGSKDT+H+++ G VD+TKPRRRRFSIS
Sbjct: 380 GKLNPFKNGSKDTSHIDEDLGGVDITKPRRRRFSIS 415
>gb|AAK59568.1| unknown protein [Arabidopsis thaliana] gi|15810641|gb|AAL07245.1|
unknown protein [Arabidopsis thaliana]
gi|23397072|gb|AAN31821.1| unknown protein [Arabidopsis
thaliana]
Length = 593
Score = 169 bits (427), Expect = 3e-41
Identities = 99/156 (63%), Positives = 114/156 (72%), Gaps = 23/156 (14%)
Frame = -1
Query: 611 LKLSYEARVHASQNKRLPVQIVLHALYYDQLRLRSGADDG--------GEAVAEREQLQT 456
LKLSY+AR+HASQNKRLPV IVLHALYYDQL+LRSG + EA+ R QLQ
Sbjct: 438 LKLSYDARLHASQNKRLPVNIVLHALYYDQLKLRSGVAEQEERAVVVLPEALKTRSQLQA 497
Query: 455 DVTLVRENEELRLELMRMKTVVSDLQK-----GVAPS------SSKKIK--FFSSVSKKL 315
D TL +ENE LR ELM+MK VSD+QK G + S SSKK K FFSSVSKKL
Sbjct: 498 DTTLAKENEALRSELMKMKMYVSDMQKNKNGAGASSSNSSSLVSSKKSKHTFFSSVSKKL 557
Query: 314 GKLNPFRNGSKDTTHLED--GPVDLTKPRRRRFSIS 213
GKLNPF+NGSKDT+H+++ G VD+TKPRRRRFSIS
Sbjct: 558 GKLNPFKNGSKDTSHIDEDLGGVDITKPRRRRFSIS 593
>gb|AAF33112.1|AF181683_1 RPT2 [Arabidopsis thaliana]
Length = 593
Score = 167 bits (424), Expect = 8e-41
Identities = 98/156 (62%), Positives = 114/156 (72%), Gaps = 23/156 (14%)
Frame = -1
Query: 611 LKLSYEARVHASQNKRLPVQIVLHALYYDQLRLRSGADDG--------GEAVAEREQLQT 456
LKLSY+AR+HASQNKRLPV IVLHALYYDQL+LRSG + EA+ R QLQ
Sbjct: 438 LKLSYDARLHASQNKRLPVNIVLHALYYDQLKLRSGVAEQEERAVVVLPEALKTRSQLQA 497
Query: 455 DVTLVRENEELRLELMRMKTVVSDLQK-----GVAPS------SSKKIK--FFSSVSKKL 315
D TL +ENE LR ELM+MK VSD+QK G + S SSKK K FFSSVS+KL
Sbjct: 498 DTTLAKENEALRSELMKMKMYVSDMQKNKNGAGASSSNSSSLVSSKKSKHTFFSSVSRKL 557
Query: 314 GKLNPFRNGSKDTTHLED--GPVDLTKPRRRRFSIS 213
GKLNPF+NGSKDT+H+++ G VD+TKPRRRRFSIS
Sbjct: 558 GKLNPFKNGSKDTSHIDEDLGGVDITKPRRRRFSIS 593
>gb|AAO16688.1| RPT2-like protein [Sorghum bicolor]
Length = 665
Score = 86.3 bits (212), Expect = 3e-16
Identities = 53/102 (51%), Positives = 63/102 (60%), Gaps = 16/102 (15%)
Frame = -1
Query: 611 LKLSYEARVHASQNKRLPVQIVLHALYYDQLRLRSGADDG---GEAVAE----------- 474
LKLSY+AR+HASQNKRLP+Q VL ALYYDQL+LRS AD G+ VA+
Sbjct: 459 LKLSYQARLHASQNKRLPLQAVLSALYYDQLKLRSAADAATSMGDVVADTQTQSSSAAGR 518
Query: 473 -REQLQTDVTLVRENEELRLELMRMKTVVS-DLQKGVAPSSS 354
R Q D L RENE LR+EL RM+ +S +Q G SS
Sbjct: 519 ARAQAMADAALARENEALRVELARMRAYMSGTMQHGKGRESS 560
>gb|AAN77296.1| Hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 626
Score = 75.9 bits (185), Expect = 4e-13
Identities = 64/171 (37%), Positives = 83/171 (48%), Gaps = 39/171 (22%)
Frame = -1
Query: 608 KLSYEARVHASQNKRLPVQIVLHALYYDQLRLRSGADD--------GGEAVA-------- 477
KLS EA HA+QN RLPVQ V+ LY++Q RLR+ + GGE+ A
Sbjct: 460 KLSREACAHAAQNDRLPVQTVVQVLYHEQRRLRAPSQPPSAAPSYAGGESPALSYRPTPS 519
Query: 476 ----EREQLQTDVT-LVRENEELRLELMRMKTVVSD-----LQKGVAPSSSKKIK----- 342
R + +V+ L REN+ELR+EL+RMK + D GV PS +
Sbjct: 520 FNGRHRPGVPDEVSRLQRENDELRMELLRMKMRLRDPSVAFSAGGVPPSGRPPLPKKPGG 579
Query: 341 --------FFSSVSKKLGKLNPFRNGSKDTTHLEDGPVDLTKPRRRRFSIS 213
F +S+SKKLG+LNPF L G V P+ RR SIS
Sbjct: 580 GGGGSSGGFMNSMSKKLGRLNPFLRSDV----LGGGRVRTKPPKDRRHSIS 626
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 525,434,985
Number of Sequences: 1393205
Number of extensions: 12224966
Number of successful extensions: 126412
Number of sequences better than 10.0: 2194
Number of HSP's better than 10.0 without gapping: 74850
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 111369
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 24568846532
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)